Team:Newcastle/Initial filamentous
From 2010.igem.org
Revision as of 23:14, 25 October 2010 by RachelBoyd (Talk | contribs)

| |||||||||||||
| |||||||||||||
Initial research: Filamentous Cells
We have chosen to produce a filamentous cell phenotype to strengthen the cracks found in concrete. Our Initial research lead us to look at the genes and systems listed below. We eventually decided on overexpressing the yneA because we think it will produce the best phenotype and only requires we control one gene. Another advantage is its small gene size.
Filamentous cells genes list
- yneA (Transcribed with yneB, ynzC, is an analogue of sulA in E.coli) * our biobrick is designed to over express this gene reducing cell division possibly by inhibiting FtsZ ring formation or constriction.
- dinR (Homologue of lexA in E.coli transcribed in the opposite direction)
- ftsZ (Involved in the recruitment of other proteins to the divisisome for cytokinesis, strangely over expression results in disruption of Zring formation as well as reduced expression)
- secA (Involved in the secretion of extracellular proteins and the insertion of transmembrane proteins)
- recA (Involved in SOS response removing the repressor DinR (LexA))
- wpr and epr produce extracellular proteases that cleave the signal peptide/transmembrane domain of YneA
- ezr produces a protein which sequesters FtsZ monomer by binding its C terminal domain and also inhibits GTP binding; however overexpression does not result in filamentation.
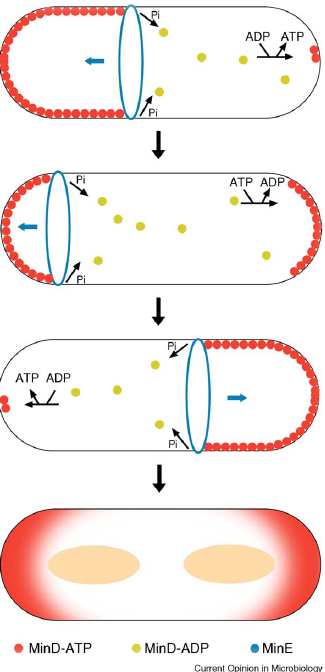
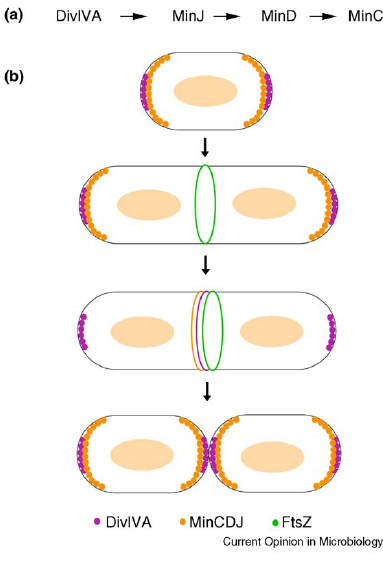
- min C,D ,J and divIVA prevent polar cell division .
- Positive regulators of FtsZ: ftsA, zapA, zipA, ftsL and divIC
- Inhibitors of Daughter cell separation: lytC,D,E,F and cwlS *Chains rather than filaments, yneA is also reported to increase the time spent in chains well into the stationary phase of bacterial growth.
 "
"