Team:SDU-Denmark/labnotes2
From 2010.igem.org
Lab notes (7/19 - 7/25)
Group: Flagella
Annealing of the two mutated strands of FlhDC (FlhDCmut)
Experiment done by: Sheila
Date: July 19th
Protocol: CP1.1
Method: PCR of the two mutated strands of the FlhDC operon
Notes: To samples were run at two different temperatures: 56,1˚C and 64,5˚C respectively.
Polymerase used: Pfu
Primers used: None, as the two strands are supposed to anneal to each other
| Pfu buffer |
5,0uL |
| dNTP |
1,5uL |
| FlhDC mutated long
strand |
1,5uL |
| FlhDC mutated
short strand |
1,5uL |
| H2O |
41,0uL |
| Pfu |
1,0uL |
The samples were run in the following PCR program:
| Program |
1st sample |
2nd sample |
Time |
| 1: Start |
94˚C |
94˚C |
3min |
| 2: Denaturing |
94˚C |
94˚C |
2min |
| 3: Annealing |
56,1˚C |
64,5˚C |
30sec |
| 4: Elongation |
72˚C |
72˚C |
2min |
| 5: |
GOTO |
Repeat |
X29 |
| 6: End |
72˚C |
72˚C |
2min |
| 7: Hold |
4˚C |
4˚C |
Results: A gel, containing the PCR product was run, using [http://www.fermentas.de/product_info.php?info=p1110 GeneRuler(TM) DNA Ladder Mix, ready-to-use (#SM0333)] loaded in lane one (from the left:
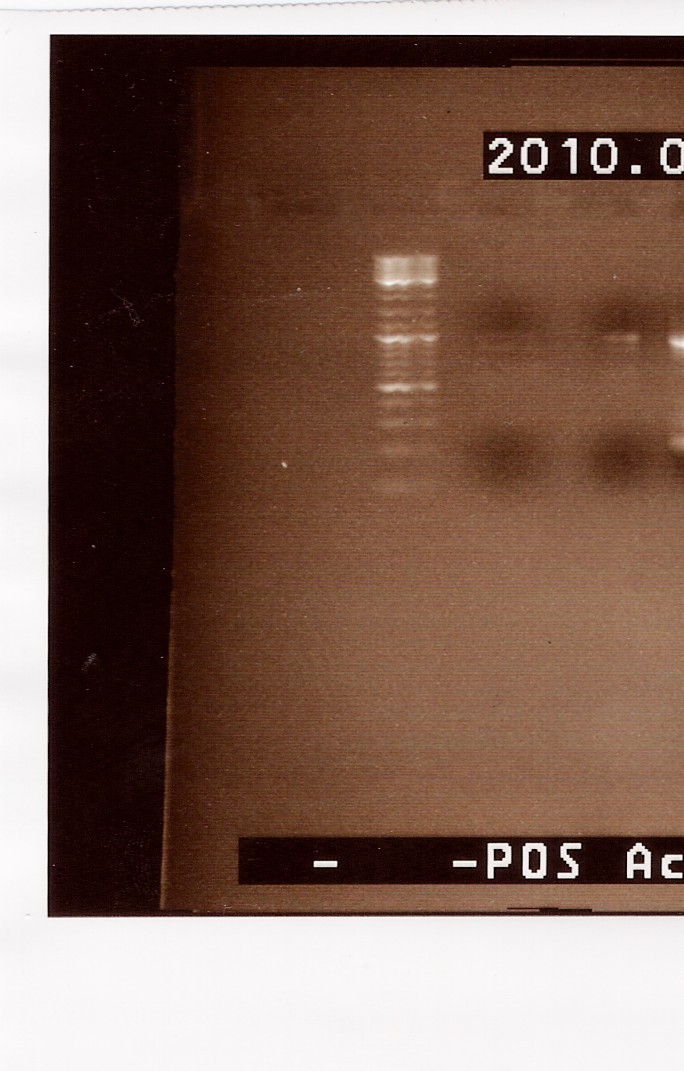
We can se very faint bands in lane 2 and 3, at around 1000bp. The band in lane 3 is a little more visible than lane 2. Because we haven't used primers ind this PCR, it is possible that the two mutated strands has a lower possibility of annealing to each other. It s however possible to amplify the few complete FlhDCmut strands in a new PCR, which is what we will do next.
--Sheila 09:10, 22 July 2010 (UTC)
Amplification of FlhDCmut
Experiment done by: Sheila
Date: July 19th
Protocol: CP1.1
Method: The experiment was done in two rounds. First, the annealing product, made by Pernille on july 19th was amplified. Then we amplified Pernilles amplification simultaneously as we amplified Sheilas previous annealing.
Primers used: FlhDC fw and FlhDC rev
Polymerase used: Pfu
Notes:
Results:
Gel extraction of FlhDCmut
Experiment done by: Sheila
Date: July 20th
Protocol:
Method:
Notes:
Results:
Restriction digest of FlhDCmut and pSB1C3 X2
Experiment done by: Sheila
Date: July 20th
Protocol: RD1.1
Method: 'Premixes:'
| FlhDCmut | X1 | X3 |
| H2O | 12µL | 36µL |
| FD Green Buffer | 2µL | 6µL |
| Enzyme A: EcoRI | 1µL | 3µL |
| Enzyme B: S | 1µL | 3µL |
| pSB1C3 | X1 | X3 |
| H2O | 12µL | 36µL |
| FD Green Buffer | 2µL | 6µL |
| Enzyme A: EcoRI | 1µL | 3µL |
| Enzyme B: X | 1µL | 3µL |
The digested samples were then run on a 1,5% agarose gel for approximately 20 minutes
Results: Because it failed the first time, this experiment was repeated, with the same results (see notes section)
The first gel
The second gel
The gels do not show any sign of a successful digest. Because the restiction enzymes only cut in the prefix and suffix of the FlhDCmut operon, it is not surprising that we do not see any bands of approximately 10bp length. However, the digestion should be very clearly visible in the plasmid backbone, as we should be cutting out RFP, which is approximately 1000bp long. The plasmid do not appear to have been digested by the restriction enzymes. 2 bands are visible, but they are both much longer than expected, in fact they appear around the length of the complete plasmid. So this experiment appears to have been unsuccessful.
Notes: First troubleshooting session led to a repeat of the experiment, as we found that human error must have been the reason for the failed results, because it was the first time i performed the experiment. However, when the repeat of the restriction digest yielded the same results, we took a closer look and found that the combination of restriction enzymes was wrong. They were the combination we will use when we begin the assembly... In this case we want to insert the biobrick (FlhDCmut) into the plasmid backbone (pSB1C3, in which case we need to use the same restriction enzymes on both the plasmid and the biobrick.
--Sheila 22:25, 1 August 2010 (UTC)
Gel extraction of FlhDCmut and pSB1C3 X2
Experiment done by: Sheila
Date: July 20th and July 21st
Protocol: DE1.3
Method: The samples were run on a 1,5% agarose gel.
Notes: No restriction were seen in the pSB1C3 lane, which lead to the conclusion that the restriction digest failed (see above. The FlhDC was extracted from the gel in order to amplify it with PCR
Results:
The first gel
The second gel
--Sheila 14:42, 2 August 2010 (UTC)
Amplification of FlhDCmut
Experiment done by: Sheila
Date: July 20th-21st
Protocol: CP1.1
Method:
Notes:
Results:
Gel extraction of FlhDCmut
Experiment done by: Sheila
Date: July 21st
Protocol:
Method:
Notes:
Results:
Gel extraction of flhD/C (native coding sequence)
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.1
Experiment done by:Maria
Notes:
70uL of flhD/C PCR product from Amplification of flhD/C was loaded onto a 1.5 agarose gel and extracted according to protocol.
DNA was eluted in 20uL elution buffer.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhDC 1 |
19.82 |
2.43 |
0.08 |
| flhDC 2 |
25.24 |
2.22 |
0.12 |
Analysis:
Nanodrop measurements indicated a possible contamination. However the DNA was pooled and used for Digestion
--Tipi 17:22, 19 July 2010 (UTC)
Digestion of flhD/C (native coding sequence) and pSB1A2 with EcoRI SpeI
Start date: 19/07 End date: 19/07
Methods: Digestion, Gel electrophoresis
Protocol:RD1.1
Experiment done by:Maria
Notes:
purified pSB1C3 (tube 18 blue) and flhD/C from Gel extraction was digested with EcoRI and SpeI.
Restriction mixture:
| flhD/C |
pSB1C3 |
|
| H2O |
30uL |
24uL |
| EcoRI |
4uL |
2uL |
| SpeI |
4uL |
2uL |
| FD green buffer |
8uL |
4uL |
| DNA |
38uL |
10uL |
| total vol. |
84 |
42 |
samples were loaded onto a 1.5 agarose gel. Gene ruler red were used as marker.
Results:
Gel electrophoresis:
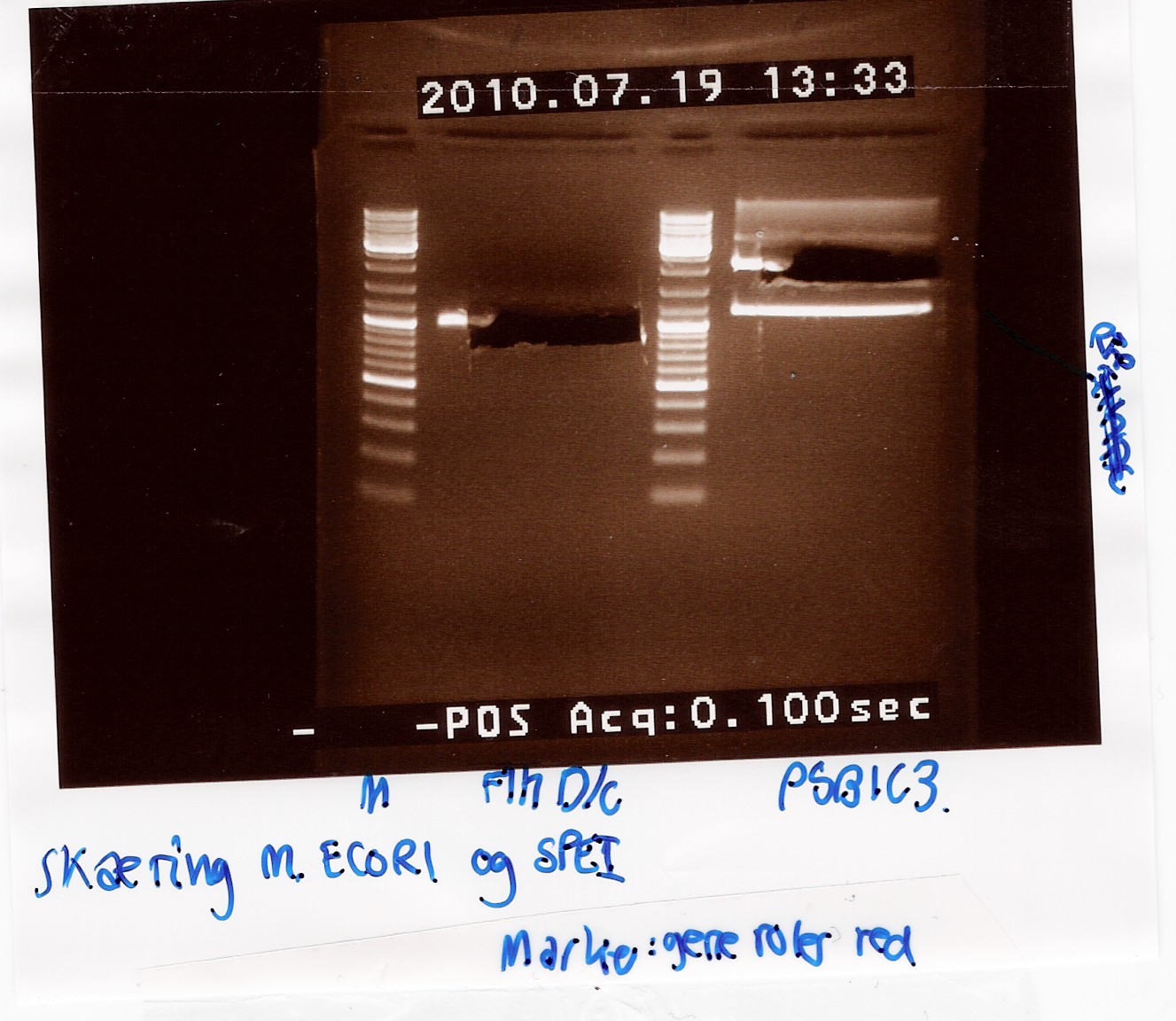
Analysis:
In lane 2 containing pSB1C3 2 bands are detected indicating a succesful digestion of the plasmid (the band at 1000 bp corresponds to J04450). A succesful digestion of the flhD/C cannot be concluded from the gel. However both bands was excised and extracted from gel (Gel extraction)
--Tipi 17:42, 19 July 2010 (UTC)
Gel extraction of digested flhD/C (native coding sequence) and pSB1C3
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.2
Experiment done by:Maria
Notes:
Digested flhD/C and pSB1C3 from Digestion were extracted from gel according to protocol.
DNA was eluted in 20uL H20.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhD/C |
18.18 |
4.59 |
0.18 |
| pSB1C3 |
14.30 |
1.89 |
0.04 |
Analysis:
nanodrop measurements indicated contamination. However both samples were used for Ligation.
--Tipi 17:57, 19 July 2010 (UTC)
Ligation of flhD/C (native coding sequence) and pSB1C3
Start date: 19/07 End date: 20/07
Methods: Ligation
Protocol:LG1.2
Experiment done by:Maria
Notes:
3 ligation reactions was prepared.
| LG1 |
LG2 |
LG3 |
|
| 10x T4 ligase buffer |
2uL |
2uL |
2uL |
| flhD/C |
5uL |
5uL |
5uL |
| pSB1C3 |
1uL |
2.8uL |
5uL |
| H20 |
11uL |
9.2uL |
7uL |
| T4 ligase |
1uL |
1uL |
1uL |
Ligation mixtures were not run on gel but were directly used for transformation
surplus ligation mixture was stored at -20 degrees as L1, L2 and L3.
--Tipi 18:15, 19 July 2010 (UTC)
Coloni PCR of flhD/C (native coding sequence) in pSB1C3
Start date: 21/07 End date: 21/07
Methods: Coloni PCR and gel electrophoresis
Protocol:CP1.2
Experiment done by: Maria and LC
Notes:
Coloni PCR was made on flhD/C in pSB1C3 from Transformation.
10 colonies from plates plated with cells transformed with L2 and L3 ligation mixtures (see Ligation).
Coloni 1-5 is taken from L2 plates
coloni 6-10 is taken from L3 plates
PCR was made with only half the amount of premix.
Premix for 12 PCR reactions:
| 10xtaq buffer |
30uL |
| MgCl2 |
12uL |
| VF2 |
12uL |
| VR |
12uL |
| dNTP |
6uL |
| H2O |
42uL |
| Total vol. |
114uL |
0.5uL Taq polymerase from ampliqon was used.
Taq PCR program:
| 1:Start |
94C |
2 min |
| 2: Denaturing |
94C |
1 min |
| 3: Annealing |
55C |
1 min |
| 4: Elongation |
72C |
1 min 30 s |
| 5: |
GO TO |
rep. 29x |
| 6: End |
72C |
3 min |
| 7: Hold |
4C |
PCR product was loaded onto a 2% agarose gel. Gene ruler DNA ladder mix (red) was used as marker.
VF2 - coding sequence (in pSB1C3) : 140bp
Coding sequence - VR (in pSB1C3) : 176bp
flhD/C : 932bp
VF2 - VR for pSB1C3 w. flhD/C :1248bp
VF2 - VR for pSB1C3 without insertion : 316bp
Results:
Gel electrophoresis:
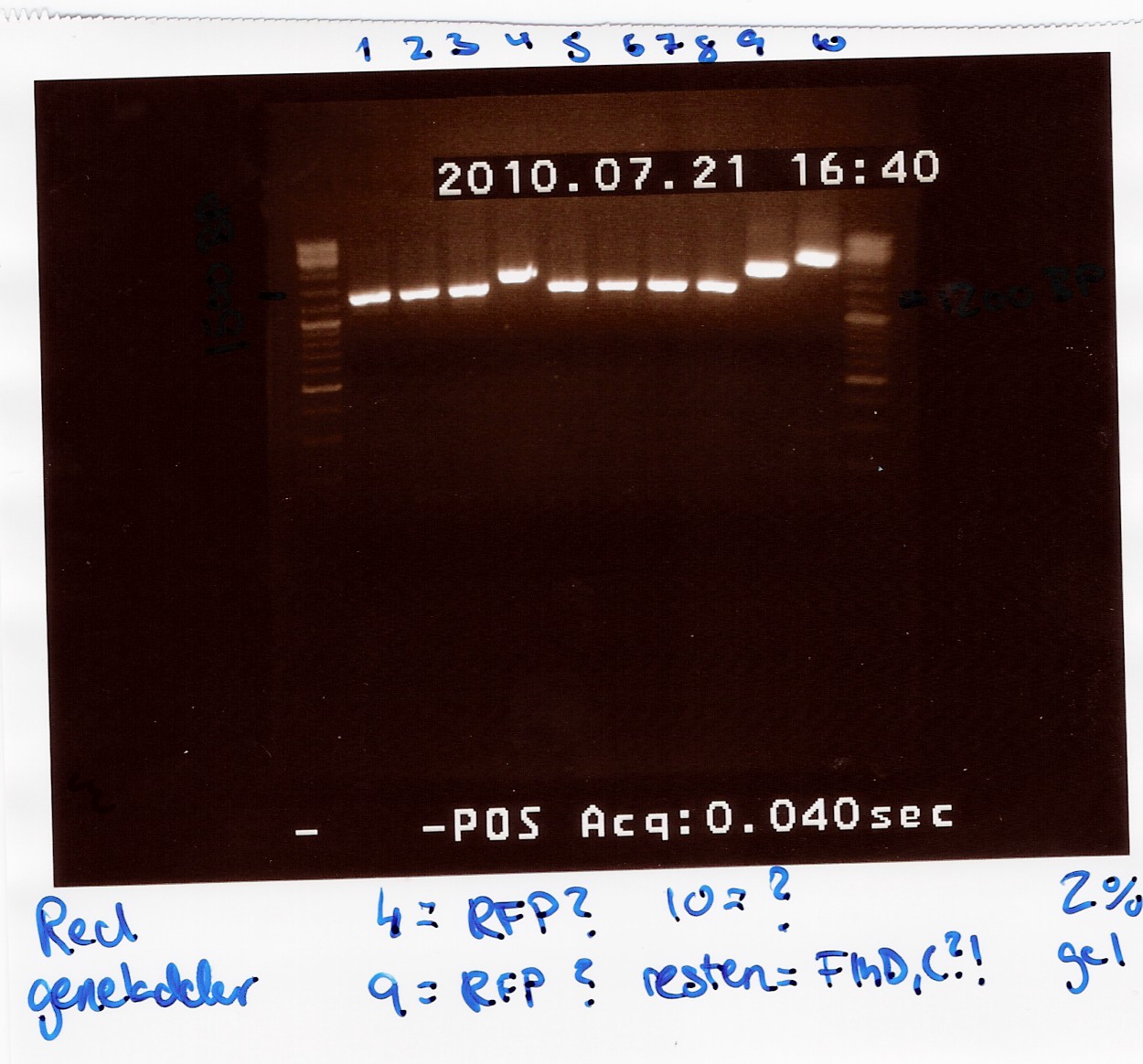
Analysis:
All lanes except 4, 9 and 10 showed a band at app. 1200bp. which might indicate plasmids where flhD/C has been inserted.
Lane 4 and 9 has a band at app. 1500bp which could indicate undigested pSB1C3 (VF2 - VR for pSB1C3 w. J04450 is 1385bp).
Streak plates of coloni 1, 2, 3, 5, 6, 7, 8 and 10 showed red colonies, indicating that these contained undigested pSB1C3.
Streak plates of coloni 4 and 9 had only white colonies, suggesting an insertion has occured.
To verify if flhD/C has been inserted coloni PCR using VF2 and flhD/C rev. as primers was carried out for colonies from streak plates of coloni 4 and 9 and of all white colonies from the transformation plates. (see coloni PCR)
--Tipi 11:46, 24 July 2010 (UTC)
Coloni PCR of flhD/C (native coding sequence) in pSB1C3
Start date: 22/07 End date: 22/07
Methods: Coloni PCR and gel electrophoresis
Protocol:CP1.3
Experiment done by: Maria
Notes:
Coloni PCR was made on flhD/C in pSB1C3 from Transformation.
All remaining white colonies from plates plated with cells transformed with L2 and L3 ligation mixtures (see Ligation) was selected.
coloni 1-6 is taken from L2.3 plates
coloni 7-15 is taken from L3.2 plates
coloni 16-27 is taken from L3.3 plates
coloni 28 is taken from streak plate of PCR no. 4
coloni 29 is taken from streak plate of PCR no. 9 (see Coloni PCR)
Premix for 31 PCR reactions:
| 10xtaq buffer |
77.5uL |
| MgCl2 |
31uL |
| VF2 |
31uL |
| flhD/C rev. |
31uL |
| dNTP |
15.5uL |
| H2O |
108.5uL |
| Total vol. |
294.5uL |
0.5uL Taq polymerase from fermentas was used.
Taq PCR program:
| 1:Start |
94C |
2 min |
| 2: Denaturing |
94C |
1 min |
| 3: Annealing |
55C |
1 min |
| 4: Elongation |
72C |
1 min 30 s |
| 5: |
GO TO |
rep. 29x |
| 6: End |
72C |
3 min |
| 7: Hold |
4C |
PCR product was loaded onto a 2% agarose gel. Gene ruler DNA ladder mix (red) was used as marker.
Analysis:
None of the samples contained amplified DNA.
This indicates either that the flhD/C gene has not been inserted to the pSB1C3 backbone or an insertion has been made but the flhD/C primer has not been able to anneal to the template.
To verify that a ligation has occurred, PCR product 4 and 9 from Coloni PCR was digested with EcoRI and SpeI.
--Tipi 13:53, 24 July 2010 (UTC)
Digestion of flhD/C PCR product with EcoRI and SpeI
Start date: 24/07 End date: 24/07
Methods: Restriction digest and gel electrophoresis
Protocol:RD1.1
Experiment done by: Maria
Notes:
To verify if a ligation has occurred Taq PCR product of coloni 4 and 9 (see Coloni PCR) was digested with the same restiction enzymes used for the digestion of flhD/C DNA and pSB1C3 prior to ligation.
If an ligation has occurred, then the restriction sites of EcoRI and SpeI should be intact, and two fragments of 122bp and 170bp should be observed on the gel, indicating VF2-E site and S site-VR respectively.
RFP (PCR poduct of coloni no.8) was used as controle.
2.5uL of undigested PCR product of coloni 4, 8 and 9 were loaded onto the gel as well.
Samples were loaded onto a 2% agarose gel. Hyper Ladder II was used as marker.
Loading scheme:
| Lane |
1 |
2 |
3 |
4 |
5 |
6 |
| digested no. 4 |
undigested no. 4 |
digested no. 9 |
undigested no. 9 |
digested no. 8 |
undigested no. 8 |
Results:
Gel electrophoresis:
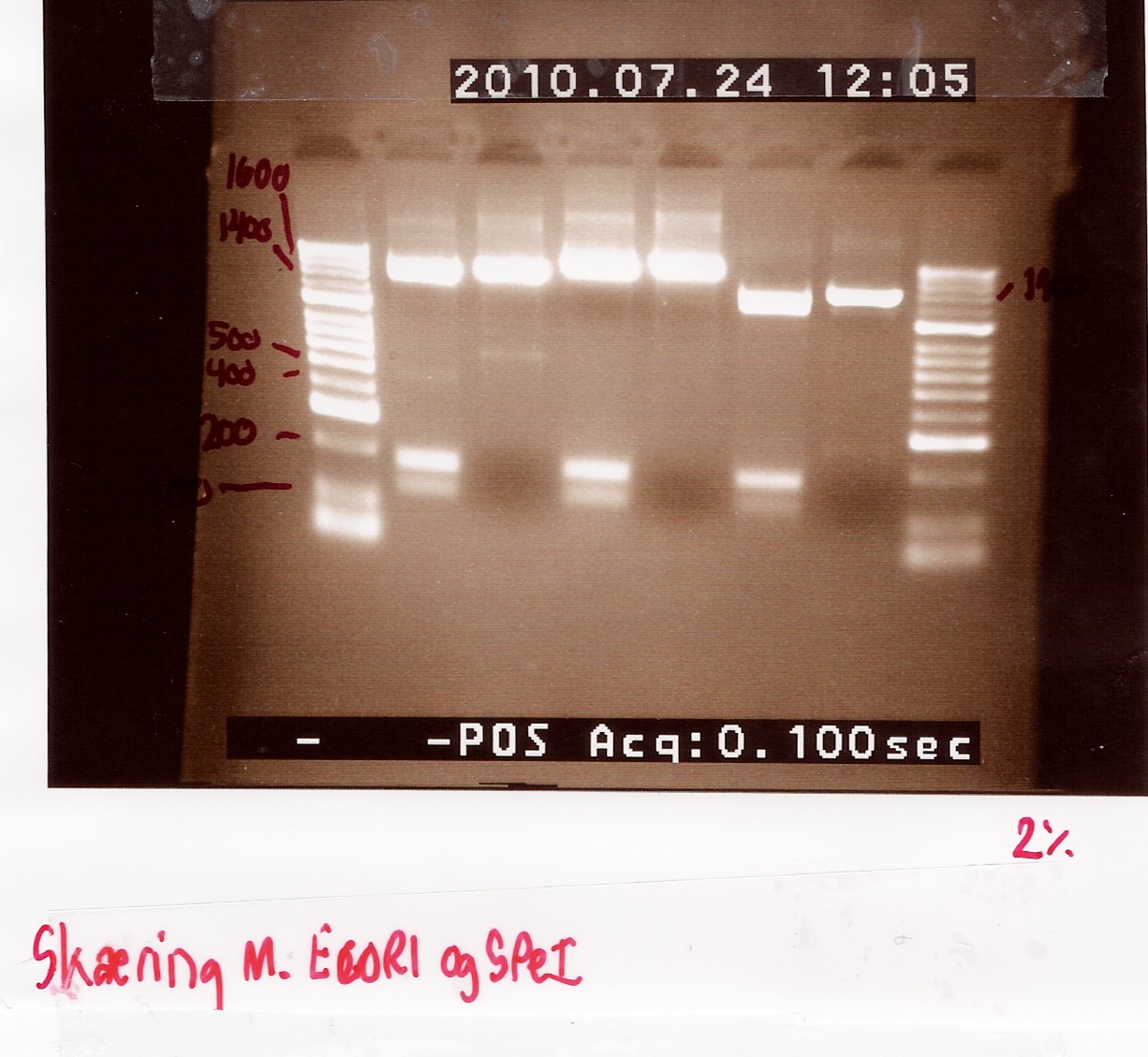
Analysis:
The 2 bands at app. 100bp and 200bp respectively indicates that the E and S restiction sites have been preserved, suggesting that a ligation has occured.
An ON culture of coloni 4 and 9 is made to use for mini-prep.
--Tipi 14:37, 24 July 2010 (UTC)
Miniprep of ligation coloni #4 and #9
Start date: 25/07 End date: 25/07
Methods: Mini prep and nanodrop
Protocol:MP1.1
Experiment done by: Maria
Notes:
The samples are loaded onto a 1.5% agarose gel. Gene ruler DNA ladder mix (red) is used as marker.
Loading sheme:
| Lanes |
1 |
2 |
3 |
4 |
| # 4.1 |
# 4.2 |
# 9.1 |
# 9.2 |
Results:
Gel electrophoresis:
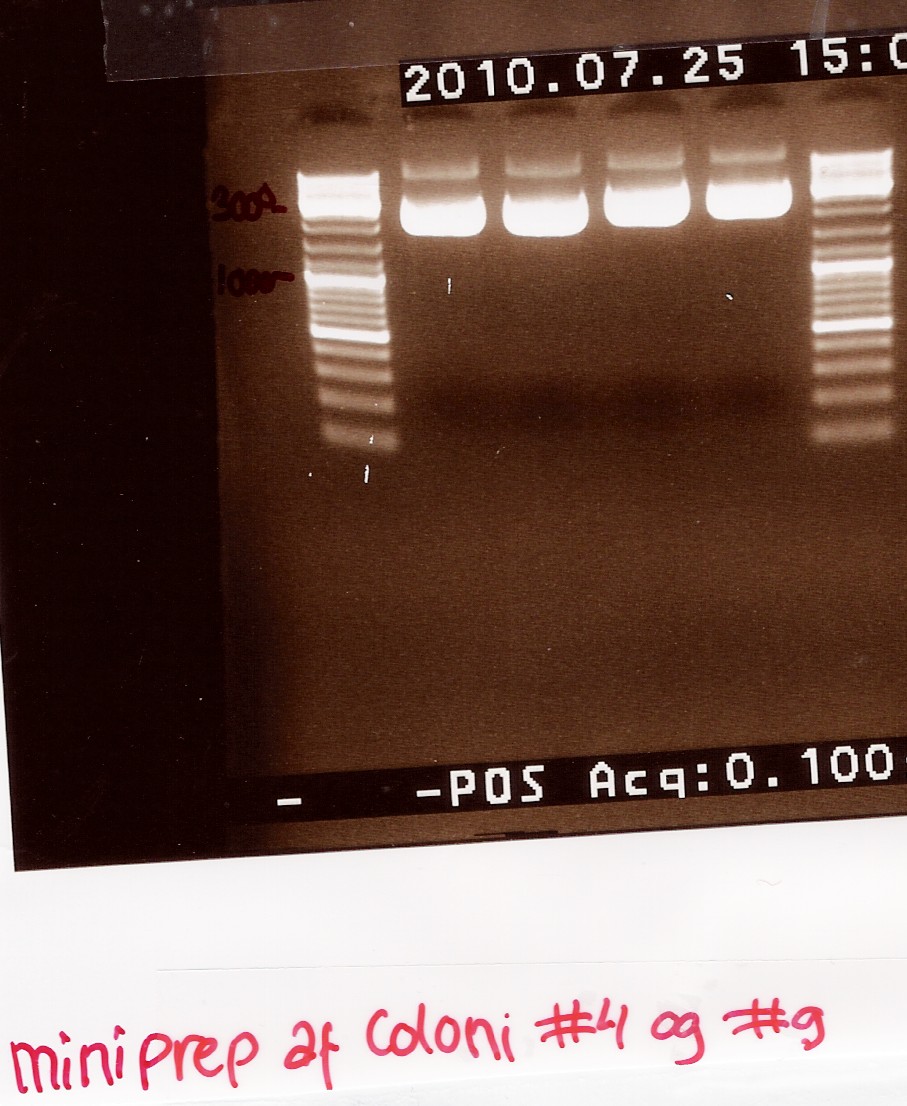
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| miniprep coloni #4.1 |
115.68 |
1.91 |
2.17 |
| miniprep coloni #4.2 |
115.74 |
1.91 |
2.13 |
| miniprep coloni #9.1 |
131.32 |
1.92 |
2.23 |
| miniprep coloni #9.2 |
119 |
1.93 |
2.16 |
| miniprep coloni #4 pooled |
116.51 |
1.92 |
2.15 |
| miniprep coloni #9 pooled |
123.94 |
1.93 |
2.17 |
Analysis:
We have purified the plasmids. The concentrations however are too low for sequencing, so the samples are dried down to reach a conc. of 200ng/uL
--Tipi 08:44, 28 July 2010 (UTC)
Digestion of flhD/C mut with PstI
Experiment no.1
Start date: 21/07 End date: 21/07
Methods: Restriction digest and gel electrophoresis
Protocol:RD1.1
Experiment done by: Maria
Notes:
To verify if the silent mutation has been made PCR product of flhD/C mut (see [flhD/C mut PCR] was digested with restiction enzyme PstI.
If the mutation has been made the flhD/C fragment should not be digested with PstI.
PCR product of flhD/C (native coding sequence) was used as controle.
Undigested PCR product of flhD/C (native coding sequence) was loaded onto the gel as well.
Samples were loaded onto a 2% agarose gel. Hyper Ladder II was used as marker.
Loading scheme:
| Lane |
1 |
2 |
3 |
| Digested flhD/C (native coding sequence) |
Digested flhD/C mut |
Undigested flhD/C (native coding sequence) |
Results:
Gel electrophoresis:
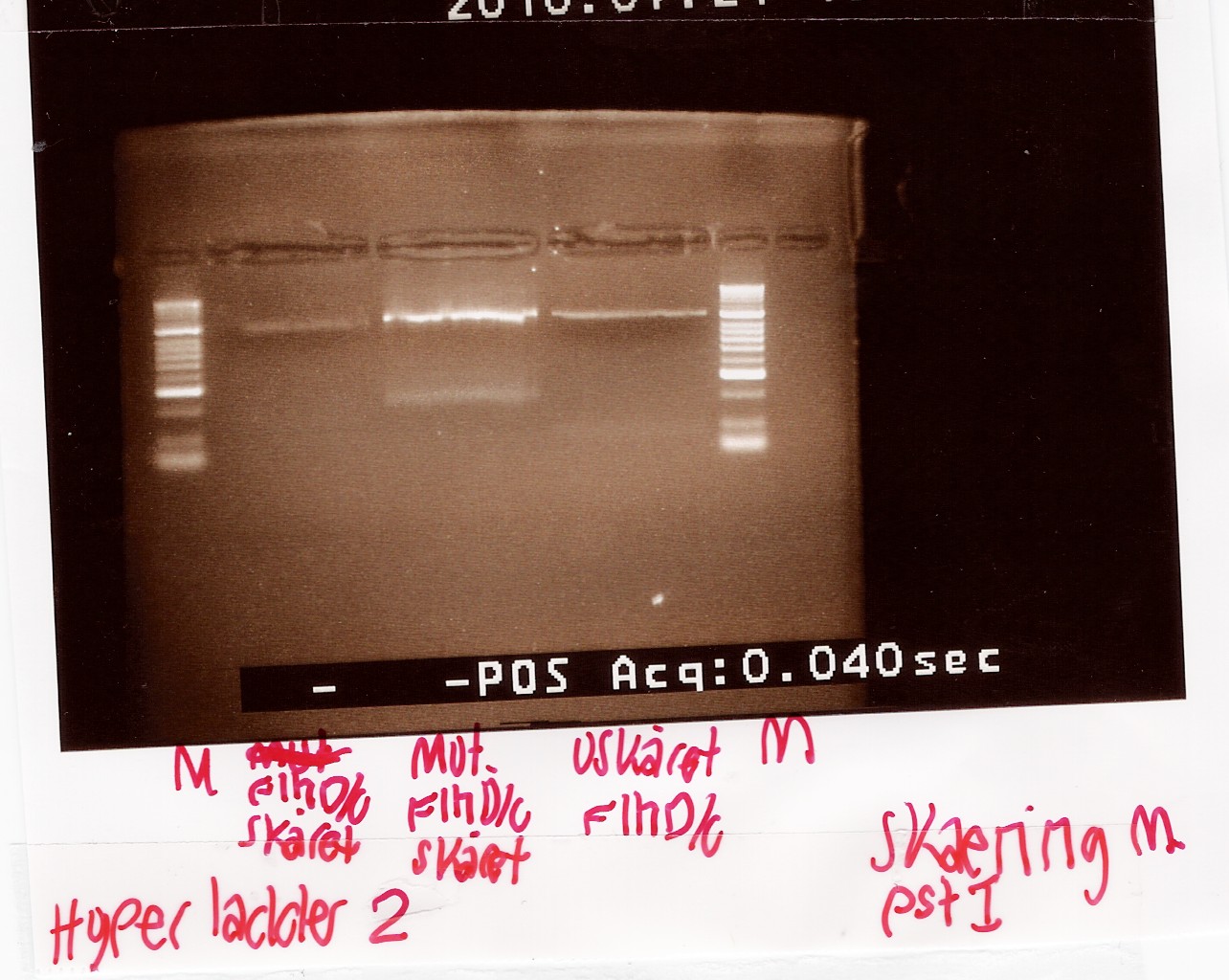
Analysis:
The intensity of the bands are too low to conclude anything. The experiment was remade using a gel with smaller wells.
--Tipi 14:37, 24 July 2010 (UTC)
experiment no.2
Start date: 22/07 End date: 22/07
Methods: Restriction digest and gel electrophoresis
Protocol:RD1.1
Experiment done by: Maria
Notes:
Taq PCR product of flhD/C (native coding sequence) (no.5 green) was used as controle.
Undigested PCR product of flhD/C (native coding sequence) was loaded onto the gel as well.
Samples were loaded onto a 2% agarose gel. Hyper Ladder II was used as marker.
Loading scheme:
| Lane |
1 |
2 |
3 |
| Digested flhD/C (native coding sequence) |
Digested flhD/C mut |
Undigested flhD/C (native coding sequence) |
Results:
Gel electrophoresis:
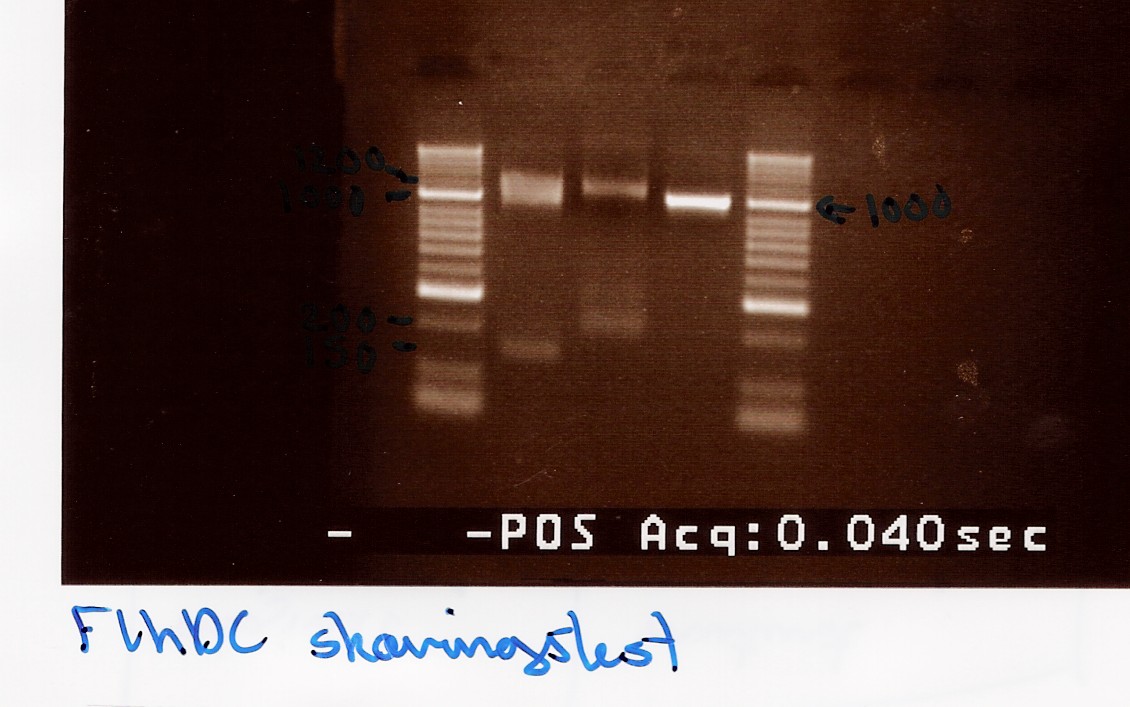
Analysis:
A small fragment is be observed for both the flhD/C mut and flhD/C (native coding sequence), so it cannot be fully concluded that the mutation has been made. Another digestion reaction must be carried out.
--Tipi 15:46, 24 July 2010 (UTC)
Purification of pSB1A2 containing J13002
Experiment done by: Pernille
Date: the 19 of July
Protocol: MP1.1 and CP1.1
Method: Miniprep, Nano drop, gel electrophoresis and PCR
Notes: The pSB1A2 plasmid are a high-copy-plasmid and therefore 5mL ON culture are used for purification of the plasmid. for the PCR the following PRC program are used:
1: 94°C 3min
2: 94°C 2min
3: 55°C 30 sec
4: 72°C 1min
5: x 29 rep.
6: 72°C 5min
7: hold 4°C
Results:After the purification the concentration of nucleic acid concentration was measured on the Nano Drop. There was made four purification tubes. Only tube 1 and 3 was used for further experiments because the other contained a very low concentration of nucleic acid. The concentration in tube 1 and 3 was 25,72ng/uL and 25,76ng/uL.
The purified DNA was loaded in a 1,5% agarose gel together with a 10.000bp long ladder (red).

PCR was run on the DNA using the VR and VF2 primers wich ensures the only the brick part are amplificated. The pfu polymerase was used but unfortunately i did not get any results since i did not add any MgSO4 to the premix.
--Pernm07 08:48, 22 July 2010 (UTC)
Amplification of pSB1A2 w. [http://partsregistry.org/Part:BBa_J13002 J13002], pSB1A2 w. [http://partsregistry.org/Part:BBa_B0015 B0015] and pSB1A2 w. [http://partsregistry.org/Part:BBa_B0034 B0034]
Experiment done by: Pernille
Date: The 20th of July
Protocol: CP1.1and DNA extraction
Method: PCR and DNA extraction
Notes: The purified DNA was amiplificated with PRC using the following program:
| Temperature (Celcius) |
Time (min) |
Description |
| 94 |
3 |
Initialization |
| 94 |
2 |
Denaturation |
| 55 |
½ |
Annealing |
| 72 |
1 |
Elongation |
| - |
x 29 |
GOTO step 2 |
| 72 |
5 |
Final Elongation |
| 4 |
|
Hold |
the experiment for each biobrick was do in duplicats.
Results:After amplification the nucleic acid concentration was measured on the Nano Drop. The concentrations was measued to:
tube 1: pSB1A2 w. J13002 was 825.2ng/uL
tube 2: pSB1A2 w. B0015 was 547.76ng/uL
tube 3: pSB1A2 w. B0034 was 777.84ng/uL
The PCR product was loaded on a 2% agarose gel together with a 1000 bp long marker.

During the DNA extraction 50uL DNA anf 7uL loading buffer was loaded in a lane. The samples was loaded from left to right in continuous numbers.

The concentration of the extracted DNA from tube 1,2 and was found to 13.60ng/uL, 16,58ng/uL and 12,6313.60ng/uL respectively. while we have to use the dobbelt terminator and the constitutive active promotor in a lot of experiment the next step is to do another round of PRC and DNA extraction on thise biobricks.
--Pernm07 10:06, 22 July 2010 (UTC)
Amplification and purification of pSB1A2 w. J13002
Experiment done by: Pernille
Date: The 22 of July
Protocol: CP1.1and DNA extraction
Method: PCR and DNA extraction
Notes: The DNA purified in the last experiment was amplified with PRC using VR og VF2 primers and pfu. the normal pfu program was used for running PCR. The PRC product was loaded on a 2% agarose gel. the brick J13002 (promotor+RBS) are 312bp long wich correspond to the visible band on the gel.

The DNA was extracted and afterwards the DNA concentration was measured on Nanodrop. the concentration was found to be 13,02ng/uL. The DNA was saved and it was cut with restriction enzymes.
Group: Photosensor
Miniprep of [http://partsregistry.org/Part:BBa_K081005 BBa_K081005] in pSB1A2 and [http://partsregistry.org/Part:BBa_R0011 BBa_R0011] in pSB1A2 from transformation 20/7
Start date: 22/07 End date: 22/07
Methods: Miniprep, gel electrophoresis and nano drop
Protocol:MP1.2
Experiment done by: LC
Notes: /
Results:
Gel electrophoresis:
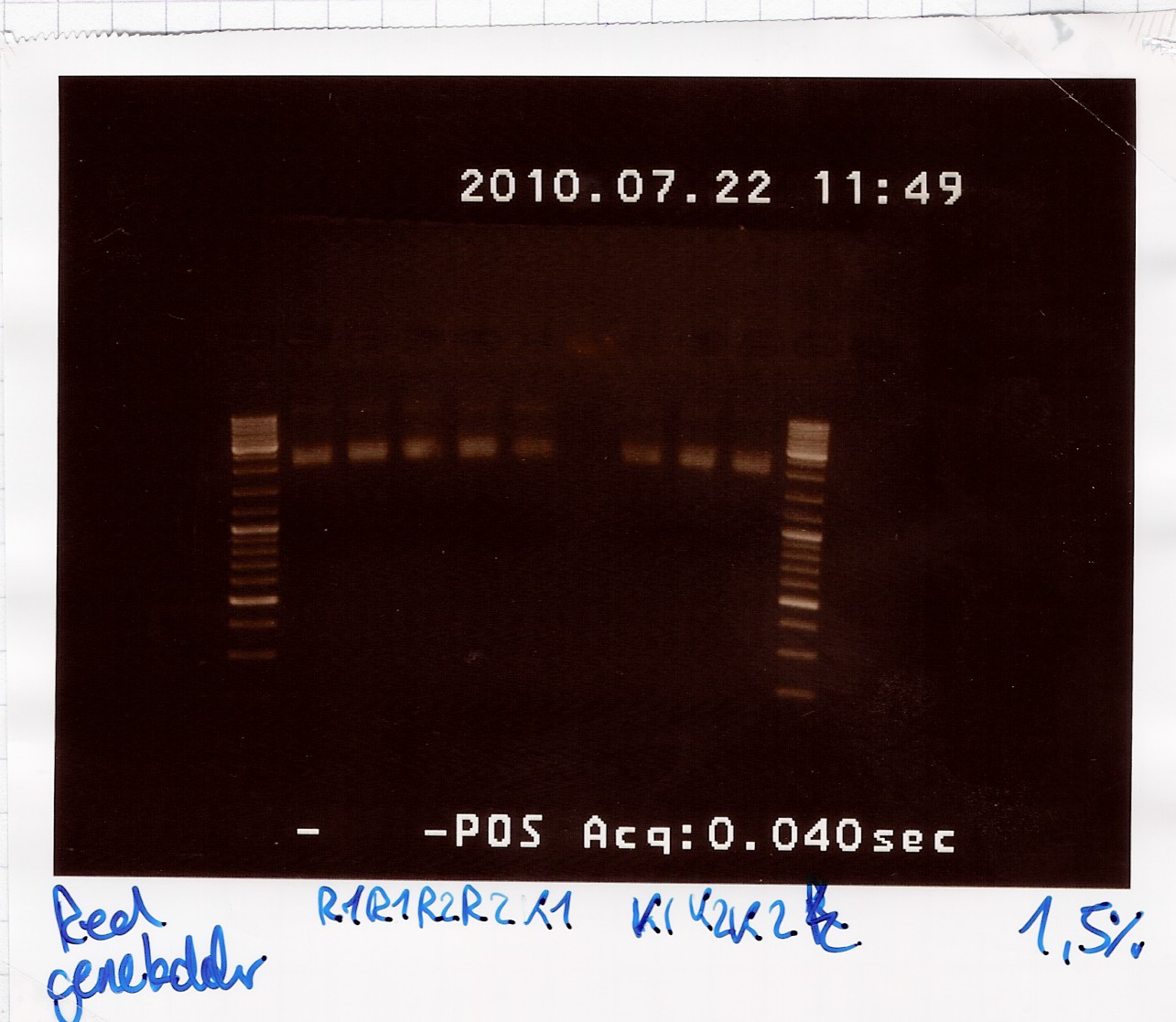
Nanodrop:
The concentration was around 23 ng/ul for all samples. Not overwhelming, but usable.
Analysis:
Sample 1-4 of BBa_K081005 were pooled and frozen and sample 1-4 of BBa_R0011 were too.
Gel extraction test (GFX vs. Fermentas)
Start date: 25/07 End date: 25/07
Methods: Gel electrophoresis, gel extraction and nano drop
Protocol:DE1.2DE1.3
Experiment done by: Maria
Notes:
FlhD/C tq PCR product (5 green) is used in the test.
PCR product is diluted in H2O to reach a sample concentration below 100ng/uL.
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| PCR product prior
to dilution |
149.93 |
0.99 |
0.38 |
| PCR product after
dilution |
89.76 |
0.99 |
0.35 |
A total of 70uL is loaded onto a 2% agarose gel.
For the GFX gel extraction 350mg of gel was used.
For the Fermentas extraction 460mg of gel was used.
Gel electrophoresis:
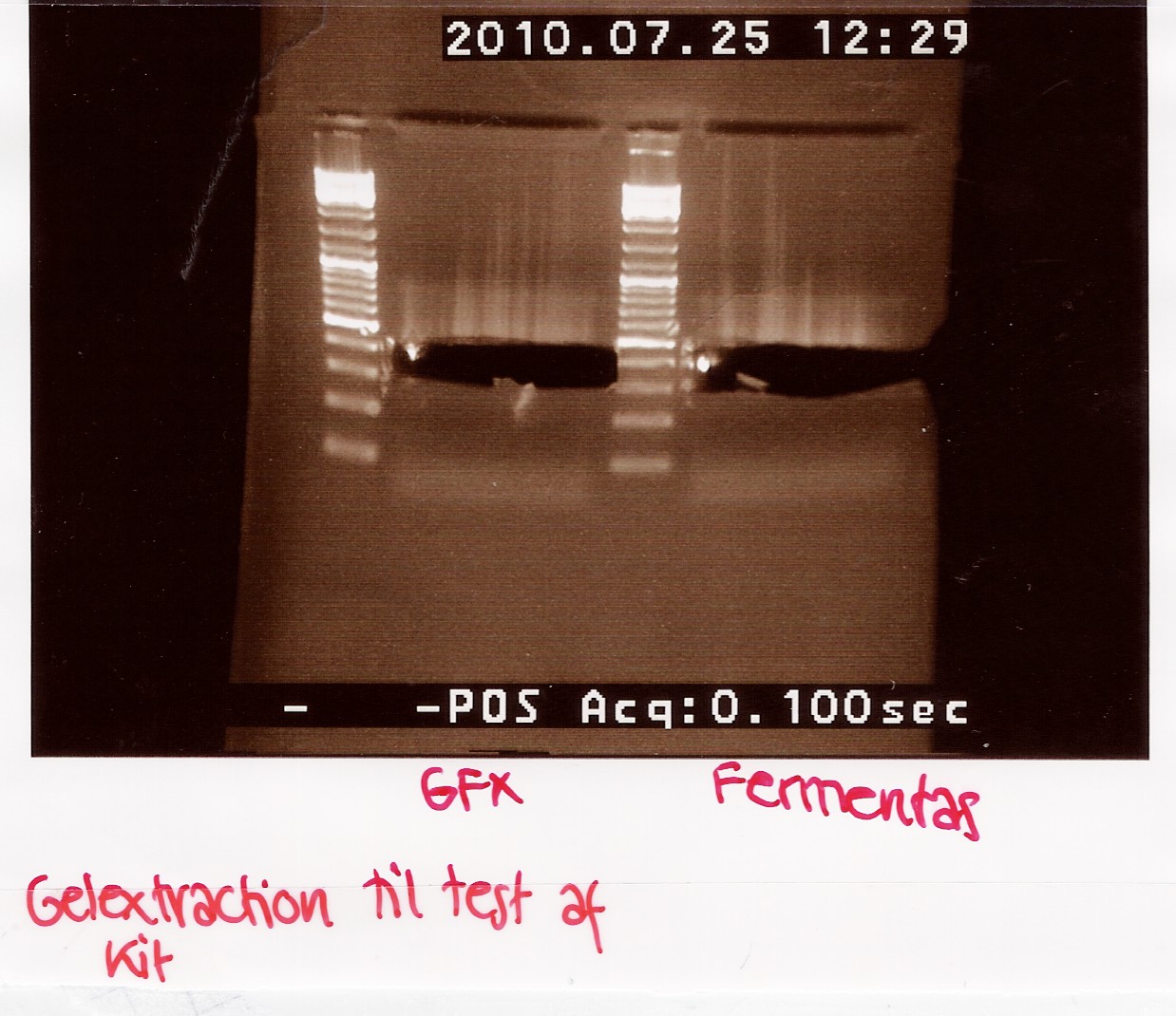
Results:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| GFX 1 |
13.72 |
2.17 |
0.08 |
| GFX2 |
16.8 |
2.02 |
0.05 |
| GFX pooled |
15.51 |
2.09 |
0.06 |
| Fermentas 1 |
20.72 |
1.9 |
0.61 |
| Fermentas 2 |
20.42 |
1.86 |
0.45 |
| Fermentas pooled |
21.38 |
1.87 |
0.51 |
Analysis:
There is extracted more DNA with the fermentas kit. This may be due to the short centrifugation time in the GFX protocol. Furthermore the GFX kit that was used was old and the buffers may have been contaminated.
Group: Retinal
Transformation of [http://partsregistry.org/Part:BBa_K081005 BBa_K081005] in pSB1A2 (constitutive promoter and RBS combined),[http://partsregistry.org/Part:BBa_R0011 BBa_R0011] in pSB1A2, pSB3C5 w. [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and pSB3T5 w. J04450 in Top 10 E.Coli
Start date: 19/07 End date: 20/07
Methods: ON culture, making competent cells, transformation
Protocol:CC1.1 TR1.1
Experiment done by: Maria
Notes:ON colony was made of 110 ml lb medium inoculated with a top10 coloni.
| Time |
Optical density |
| 8:12 |
2.9 |
| 8:17 |
0.02 |
| 9:17 |
0.035 |
| 10:17 |
0.204 |
| 10:40 |
0.380 |
| 10:50 |
0.49 |
pSB1A2 w. R0011 and pSB1A2 w. K081005 was plated with 150uL on plates containing LA, LA + Amp, LA + Tetracycline, LA + Chloramphenicol and LA + Kanamycine.Upconcentration of these samples was not made. pSB3T5 w. J04450 and pSB3C5 w. J04450 was plated according to protocol
Results:
Analysis:
Both pSB3T5 and pSB3C5 was succesfully transformed, and ON cultures with appropiate antibiotics were made for mini-prep.
For pSB1A2 w. R0011 only 6 colonies was observed on the LA+amp plate.
For pSB1A2 w. K081005 only 4 colonies was observed on the LA+amp plate.
All 10 colonies were used for Coloni PCR.
--Tipi 16:33, 19 July 2010 (UTC)
Transformation of flhD/C in pSB1C3 and test plasmid in Top 10 E.Coli
Start date: 20/07 End date: 19/07
Methods: ON culture, making competent cells, transformation
Protocol:CC1.1 TR1.1
Experiment done by: Maria
Notes:
Ligated flhD/C from Ligation and test plasmid from Whatman was transformed.
ON colony was made of 25 ml lb medium inoculated with a top10 coloni.
| Time |
Optical density |
| 8:17 |
3.5 |
| 8:20 |
0.02 |
| 9:15 |
0.035 |
| 10:30 |
0.222 |
| 10:52 |
0.350 |
| 11:05 |
0.52 |
Compotent cells were washed using 10mL 50mM CaCl2.
3 parallel transformations were carried out for L1, L2 and L3 respectively (see Ligation). L1.1, L1.2, L2.1, L2.2 and L3.1 were transformed using compotent cells from 19/7 (Compotent cells).
Prior to transformation test plasmid was washed with 10xTE pH 8.0.
Results:
Analysis:
Test plasmid was succelsfully transformed and we can carry on transforming the plasmid containing the NinaB gene.
Colonies was seen on plates transformed with L2 and L3. 10 colonies were selected and used for coloni PCR.
--Tipi 08:17, 21 July 2010 (UTC)
Coloni PCR of R0011 in pSB1A2 and K081005 in pSB1A2
Start date: 20/07 End date: 20/07
Methods: Coloni PCR and gel electrophoresis
Protocol:CP1.2
Experiment done by: Maria and LC
Notes:
6 colonies transformed with pSB1A2 w. R0011 and 4 colonies transformed with pSB1A2 w. K081005 (see Transformation) was used for coloni PCR.
Colonies with K081005 was denoted A1, A2 A3 and A4 respectively.
Colonies with R0011 was denotes B1, B2, B3, B4, B5 and B6 respectively
PCR was made with only half the amount of premix.
Premix for 12 PCR reactions:
| 10xtaq buffer |
30uL |
| MgCl2 |
12uL |
| VF2 |
12uL |
| VR |
12uL |
| dNTP |
6uL |
| H2O |
42uL |
| Total vol. |
114uL |
0.5uL Taq polymerase from ampliqon was used.
Taq PCR program:
| 1:Start |
94C |
2 min |
| 2: Denaturing |
94C |
1 min |
| 3: Annealing |
55C |
1 min |
| 4: Elongation |
72C |
30 s |
| 5: |
GO TO |
rep. 29x |
| 6: End |
72C |
3 min |
| 7: Hold |
4C |
PCR product was loaded onto a 2% agarose gel. Gene ruler 100bp DNA ladder was used as marker.
Results:
Gel electrophoresis:
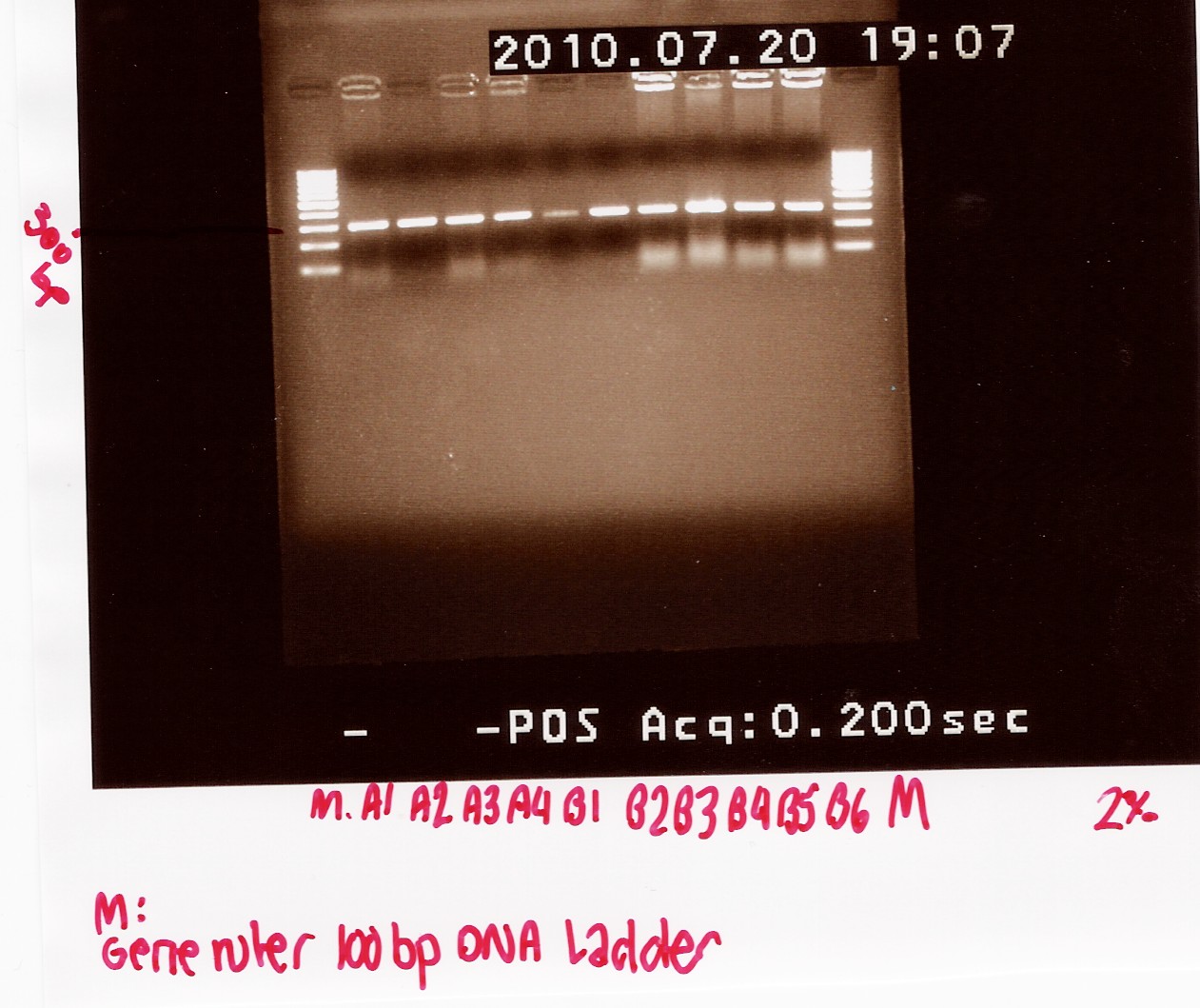
Analysis:
A band at app. 300bp was observed in all lanes, indicating that all 10 colonies contained the correct plasmids.
ON cultures were made for mini-prep.
--Tipi 06:35, 21 July 2010 (UTC)
Miniprep of pSB3T5 w. J04450 and pSB3C5 w. J04450 from transformation 20/7
Start date: 21/07 End date: 21/07
Methods: Miniprep, gel electrophoresis and nano drop
Protocol:MP1.2
Experiment done by: Maria and LC
Notes:
For pSB3T5 no.3 one half of the material was lost during the neutralization step.
pSB3T5 no.1 was eluted in 200uL elution buffer.
Samples were loaded onto a 1.5 agarose gel. Generuler DNA ladder-mix (red) was used as marker
Results:
Gel electrophoresis:
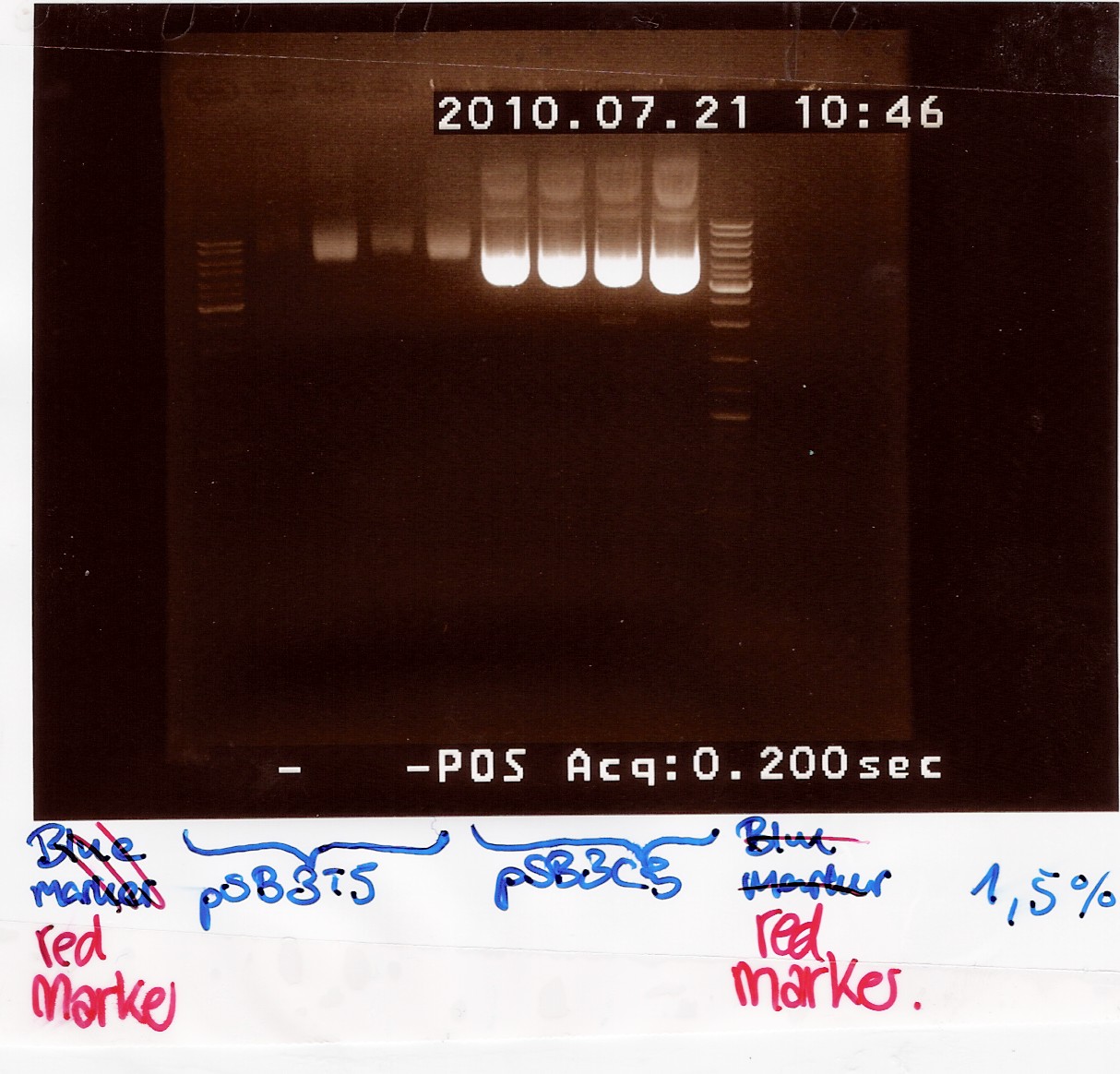
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| pSB3C5 1 |
208.2 |
1.93 |
2.31 |
| pSB3C5 2 |
225.09 |
1.93 |
2.3 |
| pSB3C5 3 |
215.2 |
1.93 |
2.3 |
| pSB3C5 4 |
260.68 |
1.93 |
2.33 |
| pSB3T5 1 |
14.16 |
2.06 |
2.0 |
| pSB3T5 2 |
52.50 |
1.96 |
2.15 |
| pSB3T5 3 |
26.45 |
1.99 |
2.3 |
| pSB3T5 4 |
53.43 |
2.0 |
2.19 |
Analysis:
Both pSB3T5 and pSB3C5 was succesfully purified.
pSB3C5 1-4 was pooled and stored as no.28 (white) at -20 degrees.
pSB3T5 1+3 was pooled and stored as no.30 (white) at -20 degrees.
pSB3T5 2+4 was pooled and stored as no.29 (white) at -20 degrees.
Nanodrop of pooled samples:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| pSB3C5 1-4 |
256.71 |
1.92 |
2.3 |
| pSB3T5 1+3 |
54.01 |
1.95 |
2.19 |
| pSB3T5 2+4 |
16.0 |
1.88 |
2.11 |
--Tipi 09:41, 21 July 2010 (UTC)
Transformation of TOP 10 e. coli with ninaB gene in pOT2:chlor vector
Date: 21/7
Done by: Christian
Transformation of cells with gene in plasmid
Date: 21/7
Methods: Transformation
Protocols: TR1.1 and paperblotter protocol from plasmid cDNA package.
Notes: Filterpaper was washed with 50ul TE buffer for 2 seconds. Paper punch was then transfered to a sterile eppendorff tube, and transformation protocol TR1.1 was followed from here. The usual positive control was used.
Plates were dried for ca. 12 minutes, showed no sign of cracking.
The gene is inserted into a pOT2 plasmid. A plasmid map can be found [http://www.fruitfly.org/about/methods/pOT2vector.html here]:
Two of the chlor plates were damaged (one very slightly) and one ampicilin plate was torn up, again. (good thing two were dried). Plates were incubated ON at 37°.
Results: Colonies showed on all plates.
Transformations have been succesfull. If our gene is in there will be tested next week.
Coloni PCR of flhD/C in pSB1C3
Start date: 21/07 End date: 21/07
Methods: Coloni PCR and gel electrophoresis
Protocol:CP1.2
Experiment done by: Maria and LC
Notes:
Coloni PCR was made on flhD/C in pSB1C3 from Transformation.
10 colonies from plates plated with cells transformed with L2 and L3 ligation mixtures (see Ligation).
Coloni 1-5 is taken from L2 plates
coloni 6-10 is taken from L3 plates
PCR was made with only half the amount of premix.
Premix for 12 PCR reactions:
| 10xtaq buffer |
30uL |
| MgCl2 |
12uL |
| VF2 |
12uL |
| VR |
12uL |
| dNTP |
6uL |
| H2O |
42uL |
| Total vol. |
114uL |
0.5uL Taq polymerase from ampliqon was used.
Taq PCR program:
| 1:Start |
94C |
2 min |
| 2: Denaturing |
94C |
1 min |
| 3: Annealing |
55C |
1 min |
| 4: Elongation |
72C |
1 min 30 s |
| 5: |
GO TO |
rep. 29x |
| 6: End |
72C |
3 min |
| 7: Hold |
4C |
PCR product was loaded onto a 2% agarose gel. Gene ruler DNA ladder mix (red) was used as marker.
VF2 - coding sequence (in pSB1C3) : 140bp
Coding sequence - VR (in pSB1C3) : 176bp
flhD/C : 932bp
VF2 - VR for pSB1C3 w. flhD/C :1248bp
VF2 - VR for pSB1C3 without insertion : 316bp
Results:
Gel electrophoresis:

Analysis:
All lanes except 4, 9 and 10 showed a band at app. 1200bp. which might indicate plasmids where flhD/C has been inserted.
Lane 4 and 9 has a band at app. 1500bp which could indicate undigested pSB1C3 (VF2 - VR for pSB1C3 w. J04450 is 1385bp).
Include column content here.
 "
"
