Team:Imperial College London/Lab Diaries/XylE team
From 2010.igem.org
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
| Team XylE |
| XylE team Lab Objectives |
|
| Lab notes and schedule | |
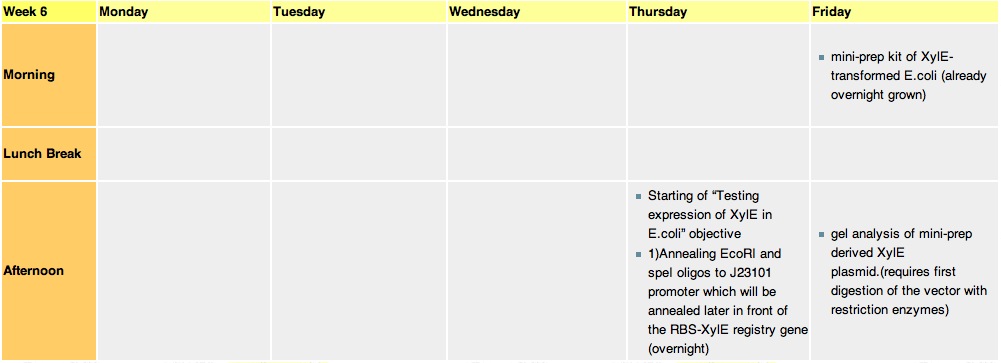
Week 6Thursday, 12-Aug-2010
we constructed the standard E.coli promoter J23101 with sticky ends. These ends are complementary to restriction sites made by EcoRI and SpeI enzyme. This promoter will be later used in 3A assemply to construct a promoter-RBS-XylE design in a psB1C3 vector. E.coli will be transformed with this final construct plasmid to assess XylE activity and characterization. It will also be one of the submitted biobricks.
these cultures are going to be used tomorrow for mini-prepping. Miniprep will allow us to isolate E.coli's plasmid DNA(which contains the XylE gene). Friday, 13-Aug-2010
Mini-prep is usually used to confirm that our gene of interest has not been changed in any way, as the isolated plasnid id sent for sequencing. However, since XylE was taken from the registry, we assume that it is fine and no sequencing is required. The mini-prep will later be used for the midi-prep (that gives out higher yeilds of DNA needed for cloning).
|
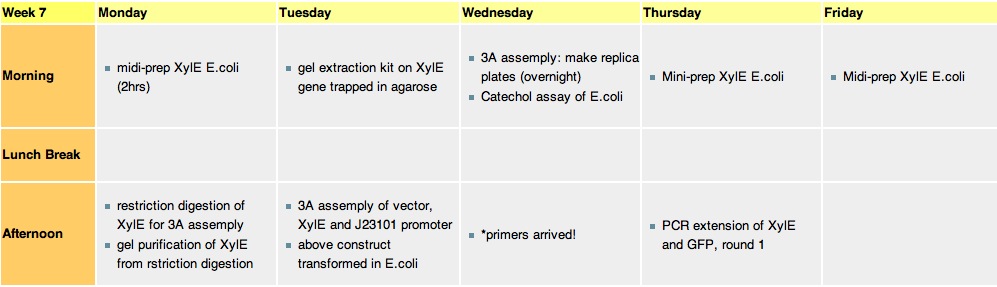
Week 7
Monday, 16-Aug-2010
- midi-prepped the XylE-transformed E.coli. The DNA yeild from the midi-prep was 134micrograms as determined by spectrophotometry. This is the XylE that is going to be used for all further experiments.
- restriction digestion of midi-prepped XylE by Xbal and PstI to prepare it for 3A assemply. (with J23101 promoter and PSB1C3 vector)
- gel analysis of the restriction digestion mixture to isolate XylE gene
Midi-prep XylE digestion with xbaI and PstI The smaller size bands at lanes 2 & 3 are the ones that are going to be cut out and used in gel purification to extract the XylE gene (with sticky ends for XbaL and PstI).
▪ I made an overnight culture of Bacillus
Tuesday, 17-Aug-2010
- using the Gel Extraction Kit, we isolated the restriction enzyme cut XylE gene from the agarose gel lamp.
- gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine their ratios for 3A assemply ligation
- 3A assemply of XylE, J23101 promoter and pSB1C3 vector.
- Transformation of XL-Blue competent E.coli with the above construct.
gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation
- I followed Chris's Bacillus transformation protocol to transform Bacillus with constitutive GFP and RFP DNA as well as a control without DNA
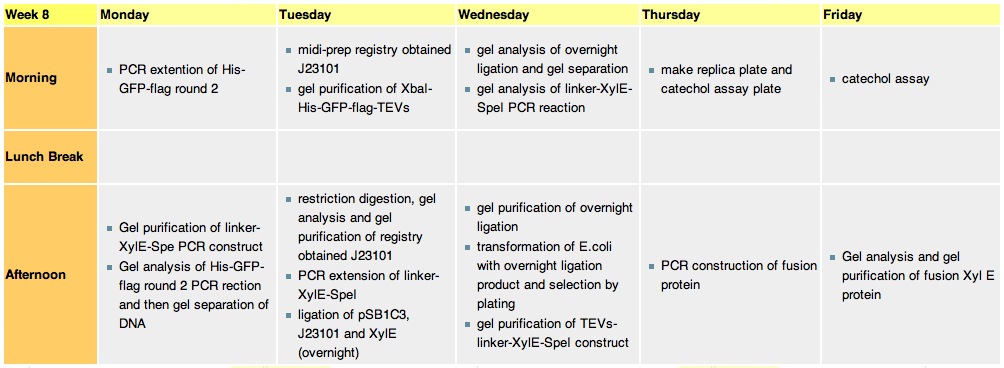
Thursday, 19-Aug-2010
- We run the XylEGFP1 PCR reaction to construct the His-GFP-Flag and Linker-XylE-Spe construct.
Friday, 20-Aug-2010
The J23101 gene in a biobrick vector containg RFP gene
- we run a gel on XylEGFP1 PCR reaction. Results: GFP was extended successfully, XylE extension FAILED (too much non-specific annealing)
- catechol assay on 2hrs bench ligation of promoter,xylE and vector failed.
- two replica plates of overnight ligated J23101, XylE and pSB1C3 transformed E.coli (one for catechol assay)
- the His-GFP-Flag DNA was gel purified
- transformation of XL-1Blue cells with J23101 in J62001 vector from the registry. One more attemp to construct a successful promoter-xylE ligation, since we believe that the strand annealed promoter was of bad quality
 "
"