Team:Newcastle/5 August 2010
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
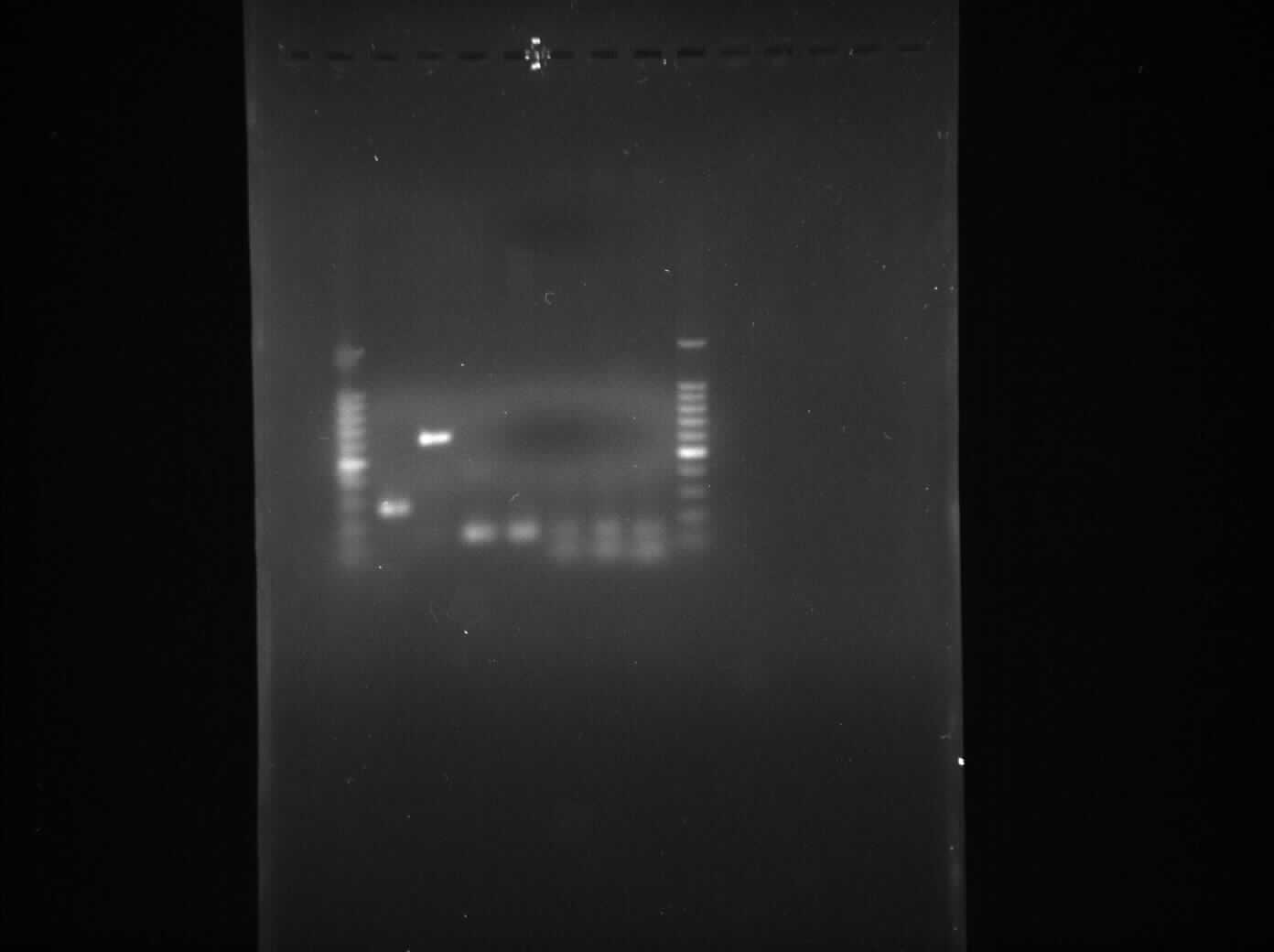
Gel Electrophoresis for the Amplified Fragments of rocF
Aim
The aim of the experiment is to perform gel electrophoresis for all the 6 PCR reactions which took place yesterday 4th August, 2010 and thus confirm that all 6 PCR reactions were successful.
Materials and Protocol
Please refer to: Gel electrophoresis.
Result
- Lane 1: 1kb DNA ladder
- Lane 2: BioBrick compatible vector pSB1C3
- Lane 3: Pspac_oid promoter
- Lane 4: 1st fragment of rocF CDS
- Lane 5: 2nd fragment of rocF CDS
- Lane 6: 3rd fragment of rocF CDS
- Lane 7: Double Terminator
- Lane 8: 1kb DNA ladder
Discussion
We found bands in the lane 2, 3, 4, 5 and 6 showing the presence of plasmid in E. coli DH5α cells. The ideal concentration of DNA calculated using nanodrop experiment is 150 µg/ml but in the table 1, where all the values have been less than 150 µg/ml which shows that even though there is plasmid present in the cells but it is present in very low amount. Also while doing nanodrop experiment, we measured 260/280 nm ratio for all the samples came out to be between 2.0 to 2.4 approximately which shows that there is a high RNA contamination in the samples.
Conclusion
This experiment shows that there is plasmid present in the E. coli DH5α cells but they are present in a very low amount and having high RNA contamination possibly due to the following reasons:
- P1 buffer which contains RNAse might be contaminated.
- RNAse enzyme might have gotten inactive.
Solution for the problem
- If P1 buffer of the Qiagen miniprep kit is contaminated, then use a different kit. We have Promega miniprep kit which will be used tomorrow.
- If RNAse enzyme is inactive, then add extra RNAse into the P1 buffer. We would be adding 10 µl in the P1 buffer solution.
 
|
 "
"