Team:Calgary/27 July 2010
From 2010.igem.org

Notebook
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
Tuesday July 27, 2010
Chris
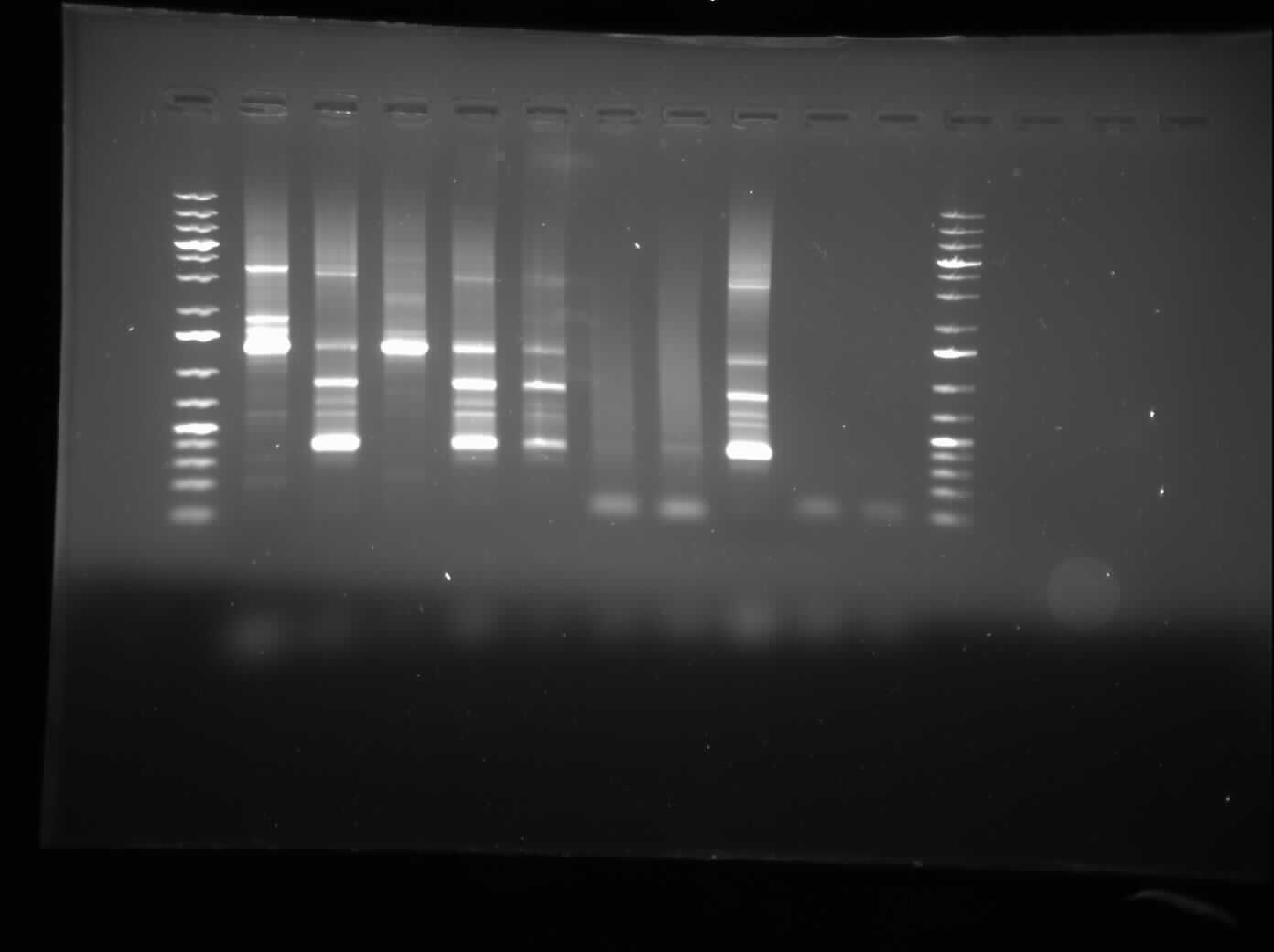
Today, I ran a 1.0% agarose gel electrophoresis of the CpxP promotor PCR that was left to be done overnight.
Jeremy
Today I ran a 0.8% gel. Prepared overnights for the I0500+B0034 in AK ccdB.
Patrick
We have decided to start using Autodesk Maya to create an animation of inclusion body formation. This means I need to learn Autodesk Maya. Thus, I spent much of the day doing so. Jeremy and I also discussed the things we need the rest of the team to help contribute to the wiki.
Emily
Today I did a gradient PCR to try to biobrick malEdelSS and malE31delSS. They are currently in the plasmids supplied to us by the Betton Labs. I also set up two more gradient PCRs to try to biobrick malE and malE31. I also set up a restriction digest with XbaI and SpeI of the PCR product from the attempt at biobricking malE and malE31 from last week. I left these to digest overnight along with psb1AC3 and psb1AK3 vectors in order to attempt to get biobricked malE and malE31 into the BBK vectors. I will transform these tomorrow and then plate them for overnight growth. I also looked into a paper on malE and the cpxP, cpxR and degP pathways.
No notebook page exists for this date. Sorry! "
"