Team:SDU-Denmark/labnotes2
From 2010.igem.org
Lab notes (7/19 - 7/25)
Contents |
Group: Flagella
Annealing of the two mutated strands of FlhDC
Experiment done by: Sheila
Date: July 19th
Protocol: CP1.1
Method: PCR of the two mutated strands of the FlhDC operon
Notes: To samples were run at two different temperatures: 56,1˚C and 64,5˚C respectively.
Polymerase used: Pfu
Primers used: None, as the two strands are supposed to anneal to each other
Results:
Gel extraction of flhD/C
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.1
Experiment done by:Maria
Notes:
70uL of flhD/C PCR product from Amplification of flhD/C was loaded onto a 1.5 agarose gel and extracted according to protocol.
DNA was eluted in 20uL elution buffer.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhD/C 1 |
19.82 |
2.43 |
0.08 |
| flhD/C 2 |
25.24 |
2.22 |
0.12 |
Analysis:
Nanodrop measurements idicated a possible contamination. However the DNA was pooled and used for Digestion
--Tipi 17:22, 19 July 2010 (UTC)
Digestion of flhD/C and pSB1A2 with EcoRI SpeI
Start date: 19/07 End date: 19/07
Methods: Digestion, Gel electrophoresis
Protocol:RD1.1
Experiment done by:Maria
Notes:
purified pSB1C3 (tube 18 blue) and flhD/C from Gel extraction was digested with EcoRI and SpeI.
Restriction mixture:
| flhD/C |
pSB1C3 |
|
| H2O |
30uL |
24uL |
| EcoRI |
4uL |
2uL |
| SpeI |
4uL |
2uL |
| FD green buffer |
8uL |
4uL |
| DNA |
38uL |
10uL |
| total vol. |
84 |
42 |
samples were loaded onto a 1.5 agarose gel. Gene ruler red were used as marker.
Results:
Gel electrophoresis:
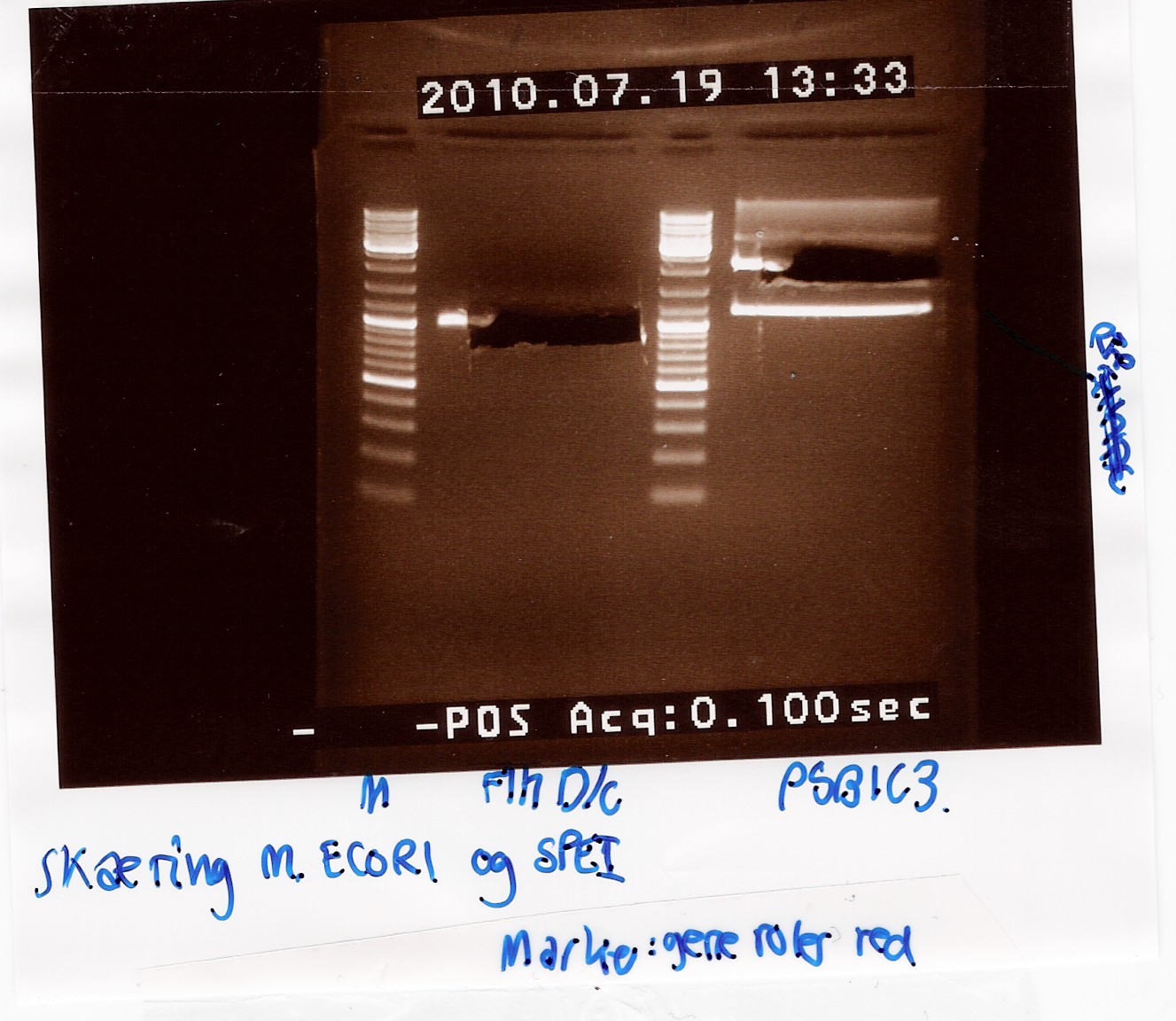
Analysis:
In lane 2 containing pSB1C3 2 bands are detected indicating a succesful digestion of the plasmid (the band at 1000 bp corresponds to J04450). A succesful digestion of the flhD/C cannot be concluded from the gel. However both bands was excised and extracted from gel (Gel extraction)
--Tipi 17:42, 19 July 2010 (UTC)
Gel extraction of digested flhD/C and pSB1C3
Start date: 19/07 End date: 19/07
Methods: Gel extraction, nanodrop
Protocol:DE1.2
Experiment done by:Maria
Notes:
Digested flhD/C and pSB1C3 from Digestion were extracted from gel according to protocol.
DNA was eluted in 20uL H20.
Results:
Nanodrop:
| Sample ID |
ng/uL |
260/280 |
260/230 |
| flhD/C |
18.18 |
4.59 |
0.18 |
| pSB1C3 |
14.30 |
1.89 |
0.04 |
Analysis:
nanodrop measurements indicated contamination. However both samples were used for Ligation.
--Tipi 17:57, 19 July 2010 (UTC)
Ligation of flhD/C and pSB1C3
Start date: 19/07 End date: 20/07
Methods: Ligation
Protocol:LG1.1
Experiment done by:Maria
Notes:
3 ligation reactions was prepared.
| LG1 |
LG2 |
LG3 |
|
| 10x T4 ligase buffer |
2uL |
2uL |
2uL |
| flhD/C |
5uL |
5uL |
5uL |
| pSB1C3 |
1uL |
2.8uL |
5uL |
| H20 |
11uL |
9.2uL |
7uL |
| T4 ligase |
1uL |
1uL |
1uL |
Ligation mixtures were not run on gel but were directly used for transformation
--Tipi 18:15, 19 July 2010 (UTC)
Group: Photosensor
Group: Retinal
Transformation of K081005 in pSB1A2 (constitutive promoter and RBS combined),R0011 in pSB1A2, pSB3C5 w. J04450 and pSB3T5 w. J04450 in Top 10 E.Coli
Start date: 19/07 End date: 20/07
Methods: ON culture, making competent cells, transformation
Protocol:CC1.1 TR1.1
Experiment done by: Maria
Notes:ON colony was made of 110 ml lb medium inoculated with a top10 coloni.
| Time |
Optical density |
| 8:12 |
2.9 |
| 8:17 |
0.02 |
| 9:17 |
0.035 |
| 10:17 |
0.204 |
| 10:40 |
0.380 |
| 10:50 |
0.49 |
pSB1A2 w. R0011 and pSB1A2 w. K081005 was plated with 150uL on plates containing LA, LA + Amp, LA + Tetracycline, LA + Chloramphenicol and LA + Kanamycine.Upconcentration of these samples was not made. pSB3T5 w. J04450 and pSB3C5 w. J04450 was plated according to protocol
Results:
Analysis:
--Tipi 16:33, 19 July 2010 (UTC)
Include column content here.
 "
"