Team:Newcastle/Urease
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
Calcium carbonate precipitation via urease expression
Bacillus subtilis produce urease, which catalyses the hydrolysis of urea into ammonium and carbonate (CO32-). Since the cell walls of the bacteria are negatively charged, they draw cations from the environment, including Ca2+, to deposit on their cell surface. The Ca2+ ions subsequently react with the CO32- ions, leading to the precipitation of CaCO3 at the cell surface.
In order for B. subtilis to fill up cracks in concrete, enhanced production of calcium carbonate must be achieved: we need to up-regulate urease production.
Previous experiments involving up-regulating ureA, ureB and ureC in B. subtilis have not lead to an increase in urease production. This could be due to yet unidentified genes that are involved in the process. Therefore, we looked for another strategy.
Flux balance analysis
In order to identify pathways which indirectly lead to urea hydrolysis we performed flux balance analysis using the [http://gcrg.ucsd.edu/Downloads/Cobra_Toolbox COBRA Matlab Toolbox] and [http://systemsbiology.ucsd.edu/In_Silico_Organisms/Other_Organisms a model of the core B. subtilis 168 metabolic network].
Flux balance analysis (FBA) is a widely used approach for studying biochemical networks. FBA calculates the flow of metabolites through a metabolic network, thereby making it possible to predict the growth rate of an organism or the rate of production of a biotechnologically important metabolite under some set conditions. [http://www.nature.com/nbt/journal/v28/n3/abs/nbt.1614.html]
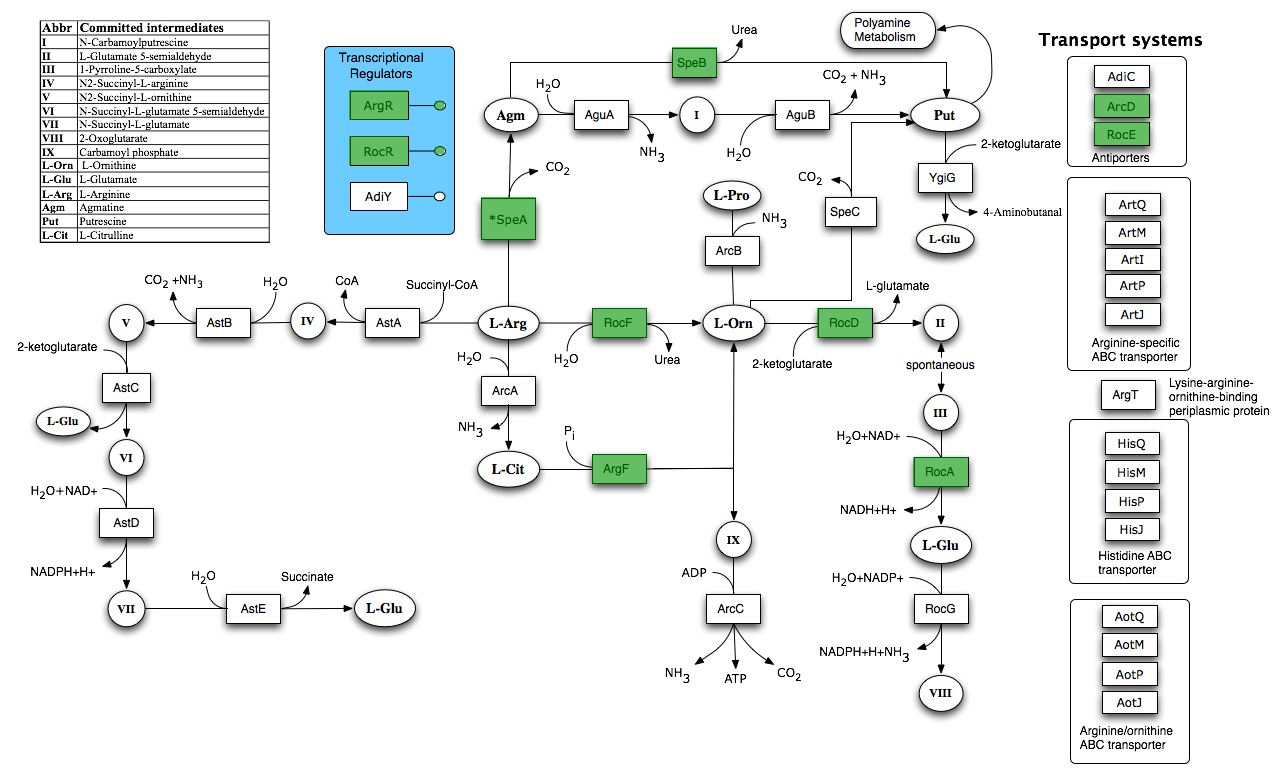
By using FBA to calculate the flow of metabolites through the B. subtilis 168 biochemical network during maximum urease activity, we were able to identify the arginine biosynthesis and catabolism pathways as potential targets.
............................................................... Results under standard conditions, opt growth ? Results under max urease activity..
Matlab file to do both
....................................................................
Parts
By increasing arginine and arginase production we can increase urea hydrolysis indirectly. Arginase breaks down arginine to urea and ornithine, leading to an increase of urea inside the cell. We believe that in turn the urea itself will increase urease production.
We plan to produce two parts, one which will enhance arginine production, and one which will enhance arginase production. These will be combined into a composite urea/urease part.
Arginine Part
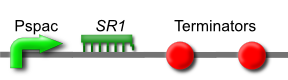
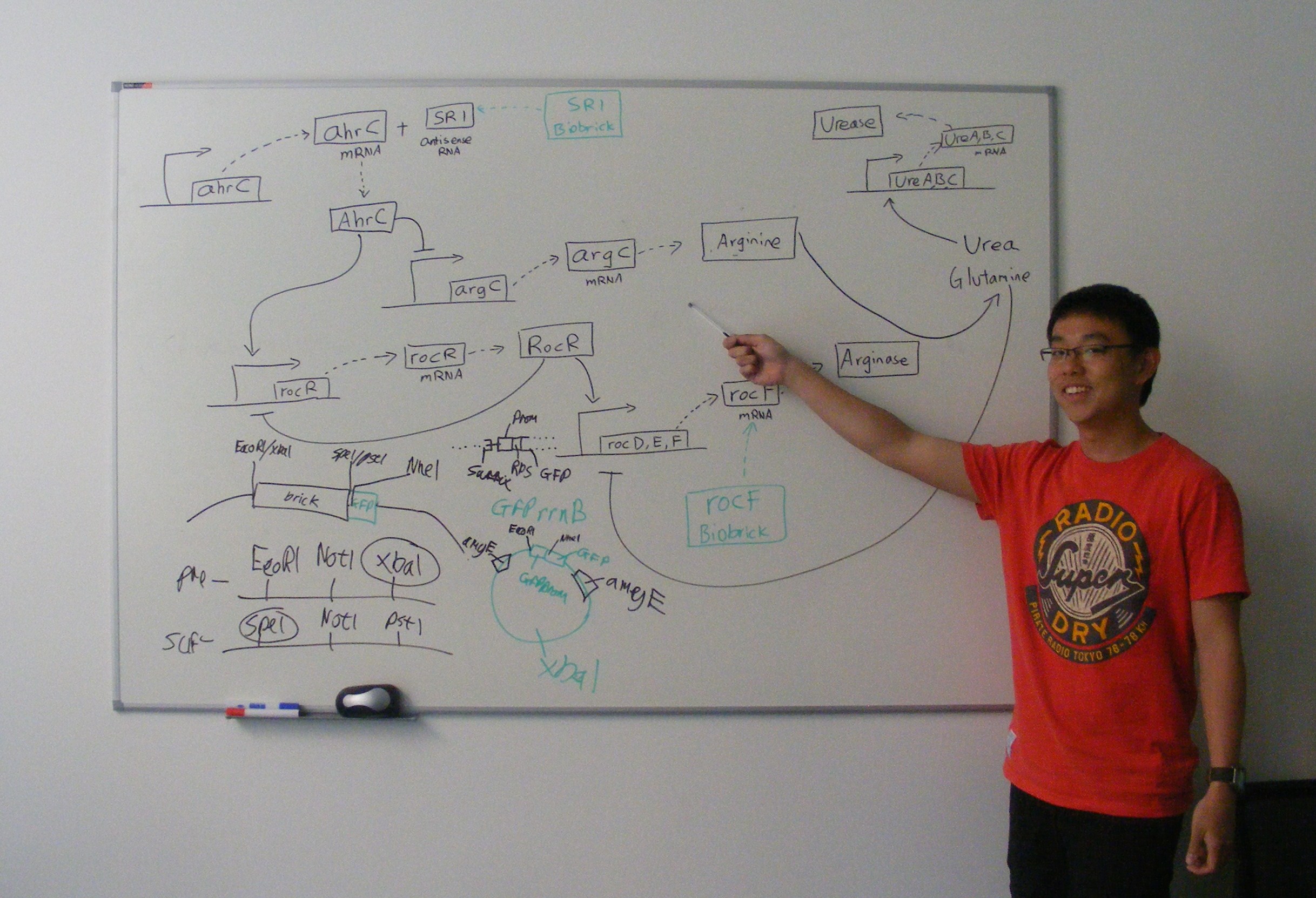
SR1 is a small untranslated regulatory RNA from the Bacillus subtilis genome. It acts as an antisense RNA to ahrC mRNA thereby inhibiting its translation. ahrC mRNA encodes the AhrC protein, which represses arginine biosynthesis and positively regulates arginine catabolism.
Transcription of SR1 results in an increase in arginine biosynthesis and a decrease in arginine catabolism, and therefore an overall increase in the level of arginine.
This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302013 part BBa_K302013] on the [http://partsregistry.org parts registry].
Arginase Part
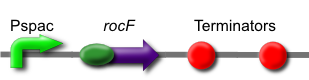
The rocF gene codes for the enzyme arginase, which breaks arginine into ornithine and urea. This is [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302014 part BBa_K302014] on the [http://partsregistry.org parts registry].
Composite urea/urease Part
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K302015 Part BBa_K302015] on the [http://partsregistry.org parts registry] combines the above two parts. The part increases urea hydrolysis indirectly, by increasing arginine and arginase production. Arginase breaks down arginine to urea and ornithine, leading to an increase of urea inside the cell. In turn the urea itself leads to urease production. Urease breaks urea into ammonia and carbonate ions and the carbonate ions are then transported to the extracellular face of the cell membrane.
Biochemical network
Taken from [http://seed-viewer.theseed.org/seedviewer.cgi?page=Subsystems&subsystem=Arginine_and_Ornithine_Degradation&organism=224308.1 SEED]
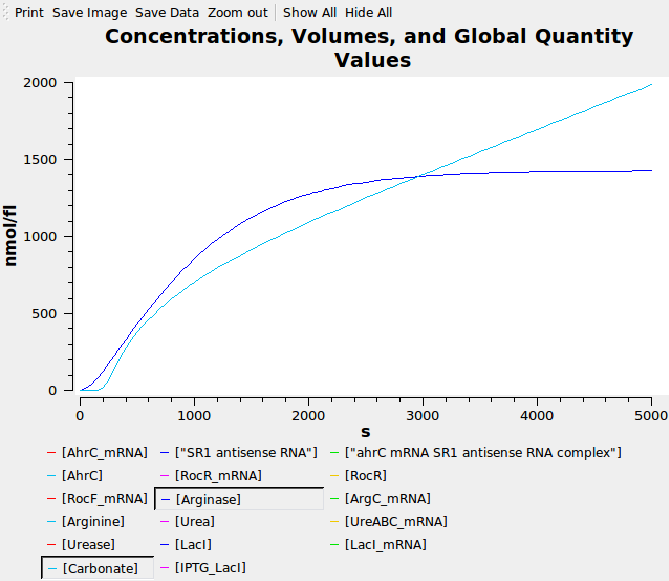
Computational model
...
Cloning strategy
- Cloning strategy for SR1
- Cloning strategy for rocF
- Cloning strategy for composite Urease BioBrick
Characterisation
See [http://www.nature.com/nbt/journal/v26/n7/abs/nbt1413.html] [http://partsregistry.org/wiki/index.php/Part_Types:Measurement_Systems] [http://partsregistry.org/Part:BBa_F2620] for more information on how to characterize a part.
References
- Jeffrey D Orth, Ines Thiele and Bernhard Ø Palsson. 2010. "What is flux balance analysis?" Nature Biotechnology. 28, p.245–248.
- Kim JK, Mulrooney SB, and Hausinger RP. 2005. "Biosynthesis of Active Bacillus subtilis Urease in the Absence of Known Urease Accessory Proteins". Journal of Bacteriology.p.7150–7154.
- Tittelboom KV, Belie ND, Muynck WD, Verstraete W. 2010. "Use of bacteria to repair cracks in concrete". Cement and Concrete Research. 40. p.157–166.
- Gardan R, Rapoport G and Debarbouille M. 1995. "Expression of the rocDEF Operon Involved in Arginine Catabolism in Bacillus subtilis". Journal of Molecular Biology. 249, p.843–856.
- Canton B, Labno A,and Endy D. 2008. "Refinement and standardization of synthetic biological parts and devices". Nature Biotechnology. 26, p.787-793.
 
|
 "
"