Team:Newcastle/Filamentous Cells
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
Filamentous cell formation via overexpression of yneA
Research
SOS response is believe to be a universal bacteria phenomenon first studied in E.coli -LexA, recA
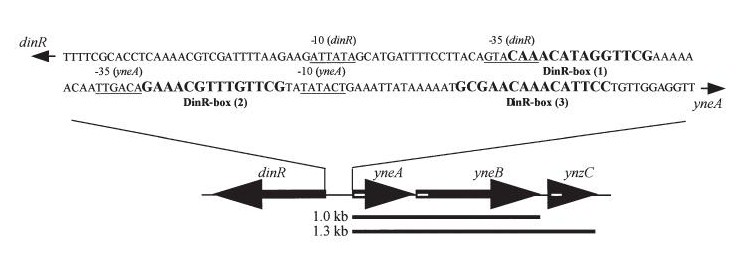
In Bacillus subtillis (gram positive) dinR protein is homologous to lexA (Repressor of din-damage inducible genes). din genes include uvrA, uvrB, dinB, dinC dinR and recA. DNA damage inhibits cell division.
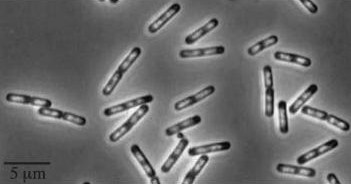
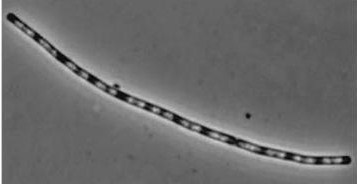
| dinR KO | Wild type Bacillus subtillis |
|---|---|
| 
|
dinR KO mutant over expressed the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (recA independent SOS response)
YneA suppressed in wt without SOS induction # Expression of YneA from IPTG controlled promoter in wt leads to elongation.
Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology.
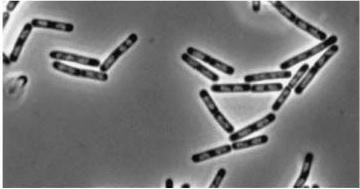
| Double mutant (dinR YneA) |
|
YneA protein required to suppress cell division. Not chromosome replication or segregation.
FtsZ is important for bacterial cell division forming a ring structure at the division site by polymerising assembling other proteins necessary for division at the site.
FtsZ localises to the cell division cycle unless dinR is disrupted or YneA is being induced. YneA suppresses FtsZ ring formation- no proven direct interaction by two-hybrid.
Filamentous cells less colony formation.
YneA expression via the inactivation of dinR by Rec A is important.
Sequence
Sequence of YneA: http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=1918391&to=1918738&report=graph&content=5
Biochemical Network
Computational Model
Cloning strategy
Media:yneA cloning strategy.pdf
 "
"