Team:Groningen/21 June 2010
From 2010.igem.org
Week 25, Arend Jan
Control restriction of pNZ8901-bbs with biobrick site enzymes. All were also cut with XhoI to check the orientation of the sites (in case the sites in the original plasmid were not removed).
- 10ul plasmid - 1.5ul buffer G (XbaI, SpeI) or R (PstI) - 0.5ul biobrick enzyme (XbaI, SpeI, or PstI) - 0.5ul XhoI - 2.5ul MQ
Because of the added XhoI a ~270bp fragment should be present. This is the case in all restrictions. All PstI restrictions give an unexpected 3 bands. The plasmid is ~800bp larger than the clone manager file but this was not considered a problem. It is likely that there is a PstI site in the unknown sequence. This site will have to be deleted before we can use this plasmid in multi-biobrick cloning steps.
As an extra control a restriction was done with BamHI which was used to linearize the plasmid after inserting the biobrick sites. Together with XhoI this should give a ~300bp band.
- 5ul plasmid - 1.5ul buffer G - 0.5ul BamHI - 0.5ul XhoI - 7.5ul MQ
300bp band is present. The biobrick sites seem to be correctly inserted. The construct will be sequenced at a later stage.
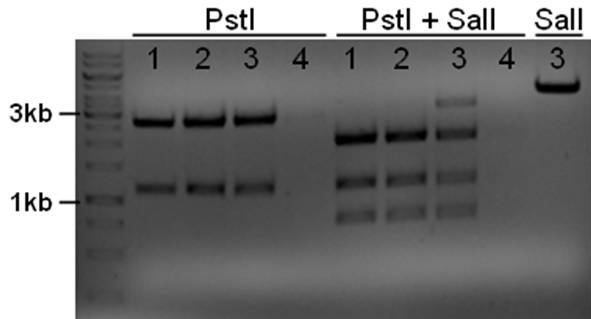
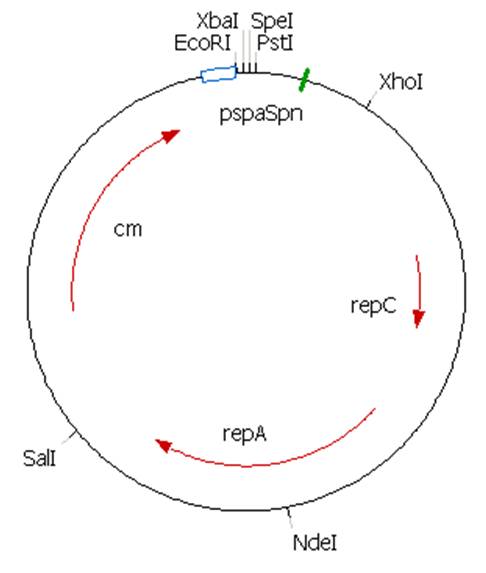
The plasmid contains an unknown sequence with an ‘illegal’ PstI site which we will need to delete. To approximate the position of the unknown sequence a restriction was performed with PstI and SalI + PstI.
- 10ul plasmid - 1.5 buffer O - 0.5/1ul enzymes (PstI or PstI + SalI(NEB)) - 3/2.5ul MQ
PstI site in unknown sequence is ~1100bp away form the PstI site in the biobrick suffix. The unknown sequence could still be either downstream of the SalI site or downstream of repC.
 "
"