Team:Groningen/21 June 2010
From 2010.igem.org
Week 25
Arend Jan
Control restriction of pNZ8901-bbs with biobrick site enzymes. All were also cut with XhoI to check the orientation of the sites (in case the sites in the original plasmid were not removed).
- 10ul plasmid - 1.5ul buffer G (XbaI, SpeI) or R (PstI) - 0.5ul biobrick enzyme (XbaI, SpeI, or PstI) - 0.5ul XhoI - 2.5ul MQ
Because of the added XhoI a ~270bp fragment should be present. This is the case in all restrictions. All PstI restrictions give an unexpected 3 bands. The plasmid is ~800bp larger than the clone manager file but this was not considered a problem. It is likely that there is a PstI site in the unknown sequence. This site will have to be deleted before we can use this plasmid in multi-biobrick cloning steps.
As an extra control a restriction was done with BamHI which was used to linearize the plasmid after inserting the biobrick sites. Together with XhoI this should give a ~300bp band.
- 5ul plasmid - 1.5ul buffer G - 0.5ul BamHI - 0.5ul XhoI - 7.5ul MQ
300bp band is present. The biobrick sites seem to be correctly inserted. The construct will be sequenced at a later stage.
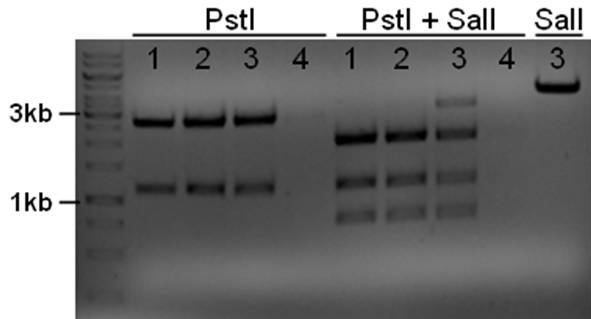
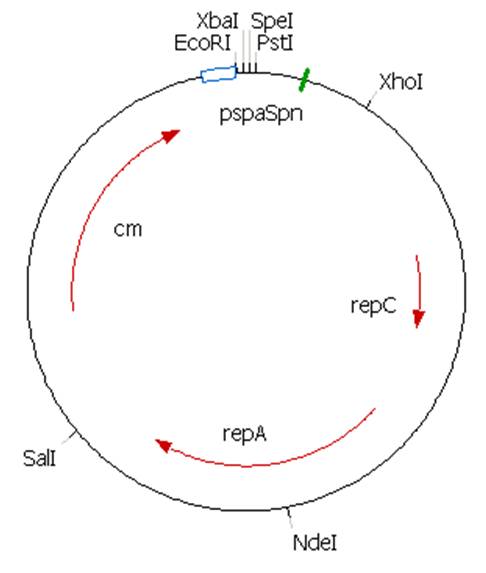
The plasmid contains an unknown sequence with an ‘illegal’ PstI site which we will need to delete. To approximate the position of the unknown sequence a restriction was performed with PstI and SalI + PstI.
- 10ul plasmid - 1.5 buffer O - 0.5/1ul enzymes (PstI or PstI + SalI(NEB)) - 3/2.5ul MQ
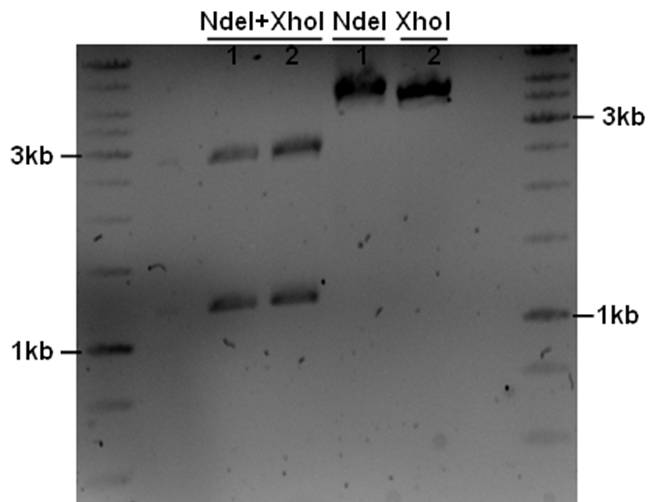
The PstI site in unknown sequence is ~1100bp away form the PstI site in the biobrick suffix. The unknown sequence could still be either downstream of the SalI site or downstream of repC. Another restriction with XhoI and NdeI will determine wether the former or the later is the case.
- 5ul plasmid - 1.5ul buffer O - 0.5/1ul enzymes - 8/7.5ul MQ
A ~1200bp fragment is present, therefore the unknown sequence must be downstream of the SalI site and upstream of the Cm gene. Sequencing primers will be designed to sequence this region.
David
Biofilm formation
Three strains of B subtilis 168 were selected for their biofilm formation abilities: - Rok - DegU - SigF - Spo0
From top to bottom: Wild-type, Rok, DegU, Spo0
.
All strains were grown overnight on either TY-agar dishes or in liquid TY-medium (TYmediumGR), 0,5% glucose was added, to test for biofilm and pellicle formation.
Inoculation with liquid culture, grown overnight:
TY-agar: 20 uL culture, glass beats. TY-agar: 5 uL culture, drop in the middle. TY-liquid medium: 5 uL
This experiment was done in duplo, growing strains at 30 and 37 degrees Celsius.
For four days, the biofilm formation was observed.
Results: Rok: Fastest growing biofilms, formed pellicle as well (Second fastest pellicle formation). DegU: Second fastest growing biofilms, formed pellicle as well (DegU grew the pellicle the fastest). SigF: Slow growing biofilm, formed pellicle as well (Showed late pellicle formation). Spo0: Slowest growing biofilm, formed a pellicle very late.
 "
"