Team:BCCS-Bristol/Modelling/BSIM/GUI
From 2010.igem.org
(→The GUI Target Market) |
|||
| Line 26: | Line 26: | ||
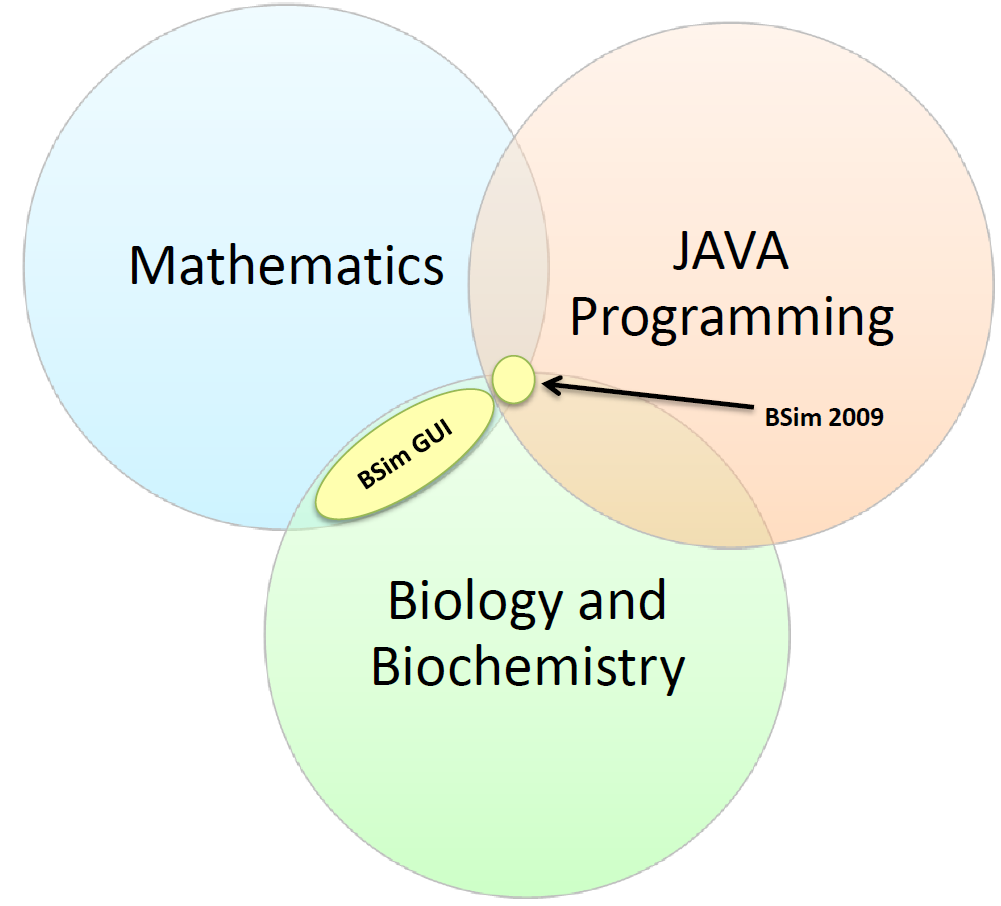
[[Image:GUI_skills_Venn_diagram.PNG|thumbnail|right|upright=3|Venn Diagram comparing the skills required to operate BSim 2009 and the new GUI]] | [[Image:GUI_skills_Venn_diagram.PNG|thumbnail|right|upright=3|Venn Diagram comparing the skills required to operate BSim 2009 and the new GUI]] | ||
| - | === | + | ===Graphical User Interface (GUI) -What's new?=== |
| - | + | Past iGEM projects at BCCS-Bristol required knowledge of JAVA, mathematics and biochemistry. The 2009 edition of BSim relied heavily on the user’s knowledge of all three. By removing the need for JAVA knowledge, we have opened up BSim to a much wider category of users. The GUI only requires users to be able to express their biological system mathematically; it can then translate the user’s description of a system into JAVA code. | |
| + | |||
| + | ===Who is the GUI aimed at?=== | ||
| + | |||
| + | The GUI is primarily aimed at users without any knowledge of JAVA. However, it is also of use to more advanced programmers, as it writes JAVA files that can be further modified using a text editor. This means advanced users can use the GUI to set a simple simulation which can be further tailored to their needs using JAVA. This saves a lot of time, as setting up a new simulation in BSim 2009 could take dozens of lines of code. | ||
Revision as of 16:48, 10 October 2010
iGEM 2010
Contents |
GUI
About the GUI - more development, how to use it is covered in 'installation'
Features
The new GUI supports many of the features of BSim 2009. These include:
- Brownian Motion
- Collisions
- Chemical fields
- Detection
- Chemical emission and consumption
- Colour signalling
- Ordinary Differential Equations (ODEs)
- Variable boundary conditions
- Directed movement (Chemotaxis)
Development
Motivation
The 2009 edition of BSim was written using an object-oriented programming language called JAVA. This was an excellent tool in which to construct BSim, as it made it very easy to produce agent-based models. However, modelling a specific system in BSim required the user to edit JAVA code. We envision a future in which BSim is a widely used and accessible modelling framework. To achieve this we needed to create an interface that allows users unfamiliar with JAVA to create simulations in BSim. This would allow BSim to be utilised by the entire synthetic biology community, rather than just specialist modellers.
Graphical User Interface (GUI) -What's new?
Past iGEM projects at BCCS-Bristol required knowledge of JAVA, mathematics and biochemistry. The 2009 edition of BSim relied heavily on the user’s knowledge of all three. By removing the need for JAVA knowledge, we have opened up BSim to a much wider category of users. The GUI only requires users to be able to express their biological system mathematically; it can then translate the user’s description of a system into JAVA code.
Who is the GUI aimed at?
The GUI is primarily aimed at users without any knowledge of JAVA. However, it is also of use to more advanced programmers, as it writes JAVA files that can be further modified using a text editor. This means advanced users can use the GUI to set a simple simulation which can be further tailored to their needs using JAVA. This saves a lot of time, as setting up a new simulation in BSim 2009 could take dozens of lines of code.
 "
"