Team:Washington/Bibliography
From 2010.igem.org
| Line 30: | Line 30: | ||
Play [http://files.agame.com/mirror/flash/p/puzzle_bobble.swf Puzzle Bobble]! | Play [http://files.agame.com/mirror/flash/p/puzzle_bobble.swf Puzzle Bobble]! | ||
Play [http://www.thepcmanwebsite.com/media/pacman_flash/pacman-flash.swf Pac Man]! | Play [http://www.thepcmanwebsite.com/media/pacman_flash/pacman-flash.swf Pac Man]! | ||
| + | |||
| + | |||
| + | |||
| + | =='''Analyzing CapD_CP'''== | ||
| + | |||
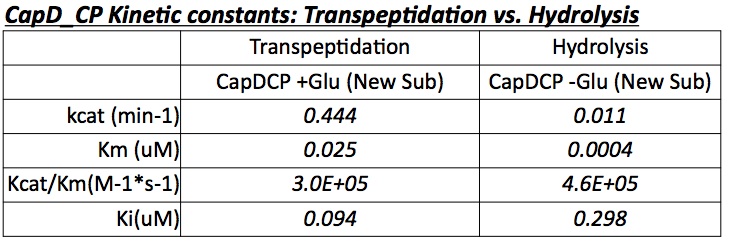
| + | [[Image:Washington_CapD_CP_Trans_Hydro_Kinectic_Table_full.jpg|thumb|400px|right|Figure 4 shows the kinetic constants of CapD_CP's transpeptidation and hydrolysis capability]] | ||
| + | |||
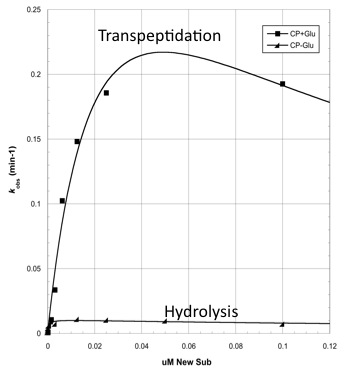
| + | [[Image:Washington_CapD_CP_Michaelis_Curve.jpg|thumb|300px|right|]] | ||
| + | <br style="clear: both" /> | ||
| + | |||
| + | The two abilities of CapD_CP are transpeptidation and hydrolysis. Based on the Kcat and Km (see figure 4) | ||
| + | values of the two, we conclude that CapD_CP is a weak binder and efficient catalyst for the transpeptidation reaction. In terms of hydrolysis, it shows strong binding but slow catalysis, thus our mutant designs focus on increasing the catalytic efficiency. | ||
| + | |||
| + | |||
| + | |||
Revision as of 18:30, 6 October 2010
Include references
Play [http://files.agame.com/mirror/flash/p/puzzle_bobble.swf Puzzle Bobble]! Play [http://www.thepcmanwebsite.com/media/pacman_flash/pacman-flash.swf Pac Man]!
Analyzing CapD_CP
The two abilities of CapD_CP are transpeptidation and hydrolysis. Based on the Kcat and Km (see figure 4) values of the two, we conclude that CapD_CP is a weak binder and efficient catalyst for the transpeptidation reaction. In terms of hydrolysis, it shows strong binding but slow catalysis, thus our mutant designs focus on increasing the catalytic efficiency.
Test: Enzyme Assay

After we have the CapD_CP mutants, we tested our mutants for their catalytic activity using our fluorescence-based enzyme assay scheme. Fluorescence-based enzyme assay measures the rate at which fluorescence in the testing media is released and the amount fluorescence depends on the rate at which fluorophore-quencher linkage is disrupted. Our substrate PDGA contains a linked fluorophore-quencher component. The faster fluorophore-quencher component is cleaved, the higher the amount of fluorescence is released and thus the greater the enzymatic activity observed.
Using Recombineering to create a constitutive T6SS
The process for using homologous recombination to replace the T6SS native promoters is simple. We begin with a previously characterised fosmid (a large, single-copy plasmid based on the F' plasmid) which contains all the genes in our T6SS. Of particular interest is the fha1-tssA1 intergenic region containing the promoters. We transform this fosmid into recombineering strain E. coli SW102, which contains heat-inducible lambda phage recombinases and lacks the galactose metabolism gene galK.
Using PCR, we create a cassette containing the galK gene flanked by 50 bp of homology to the intergenic region. The lambda recombinases integrate the cassette into the fosmid. The transformants are plated on minimal galactose media to select for the recombinants.
A second cassette is created via PCR, this time containing our new promoters. In this case, we chose to use T7 phage promoters due to their constitutive expression and small size, making them easier to synthesize. The promoters are again flanked with homologous regions, and integrated into the fosmid. This cassette displaces the galK cassette. The transformants are this time plated on 2-deoxy-galactose (DOG), which is a toxic homolog to galactose. Cells which can metabolize galactose (that is, those with a functioning galK gene) are killed, while those without the gene are unharmed.
The final step is to transform the recombinant plasmid into a T7 polymerase expression strain, to allow T6SS constitutive expression.
Overall Objective:Using Type VI Secretion as an Antibacterial Agent
Our overall goal is to clone the Type 6 secretion system and the Tse2/Tsi2 toxin/antitoxin system from Pseudomonas aeruginosa into E. coli to make a strain of E. coli that could be used to kill off gram negative pathogens present in the human gut. Ideally, this system would be regulated in such a way that the strain of E. coli would only be able to kill gram negative bacteria when a gram negative pathogen is present. This strain could (ideally) be introduced into the gut either as a preventive measure or as a treatment after a known infection. Ideally, the probiotic would only kill of gram negative bacteria in the area of infection.
Cool Picture goes here
Designing the T6SS for a E. coli probiotic
Justin and or Laura, please write this section
short intro, basic plan:
-molecular schematic of T6SS (Jesa send to Matthew)
-include map of, describe T6SS putative operon ( there is a map color coded to diagram
-mention that it is in pao1 and state objective overall: transfer into ecoli so that it functions constitutively , what genes
===Objectives:
1. figure out which genes are necessary
Mention not all genes in operon neccessarily needed for T6SS, preferable to be smaller
2. physically transfer genes into E. coli
mention large size makes it difficult, mention/explain fosmid, include fosmid map ( Laura should have This)
3. optimize regulation
mention that fosmid contains natural promoter, may not be trascribed from at all, or at right amount for a plasmid ( more than 1 copy of plasmid)
In order to create a probiotic application for this system, we first attempt to express it heterologously in non-pathogenic E. Coli. Starting from a Fosmid containing our T6SS, we are using [http://web.ncifcrf.gov/research/brb/recombineeringInformation.aspx Recombineering] to replace the strict native regulation with robust T7 promoters to create strong expression of the T6SS.
All the essential genes for our T6SS are contained within two putative operons, encoded in opposite directions. The native promoters for both operons are found in the same intergenic region, between fha1 and tssA1. Therefore, we can easily replace the promoter regions for both operons in one step.
 "
"