Team:Washington/tools/created/new-software
From 2010.igem.org
| Line 31: | Line 31: | ||
| - | ==[[Image:wikidust_banner.png|500px]]== | + | ==[[Image:wikidust_banner.png|500px|center]]== |
WikiDust is a plugin that can be used to export TinkerCell models to the iGEM wiki or other webpages. Users can then download the models for use in their own larger system, or to see how they respond when parameters are changed. We hope it will provide an easy way to share models, encouraging more teams and researchers to describe their parts quantitatively. This will allow successful reuse of their parts in the future. | WikiDust is a plugin that can be used to export TinkerCell models to the iGEM wiki or other webpages. Users can then download the models for use in their own larger system, or to see how they respond when parameters are changed. We hope it will provide an easy way to share models, encouraging more teams and researchers to describe their parts quantitatively. This will allow successful reuse of their parts in the future. | ||
| Line 76: | Line 76: | ||
<br /> | <br /> | ||
| - | ==[[Image:partsrobot_banner.png|500px]]== | + | ==[[Image:partsrobot_banner.png|500px|center]]== |
Revision as of 23:02, 5 October 2010
Software Tools
As the systems created by synthetic biologists get more complex, automation and computer-aided design will be needed. Computers will eventually play an important part in the design, construction, and testing of new devices. To meet the emerging needs of synthetic biology, the UW iGEM team has developed several new software tools.
WikiDust is a plugin that can be used to export TinkerCell models to the iGEM wiki or other webpages. Users can then download the models for use in their own larger system, or to see how they respond when parameters are changed. We hope it will provide an easy way to share models, encouraging more teams and researchers to describe their parts quantitatively. This will allow successful reuse of their parts in the future.
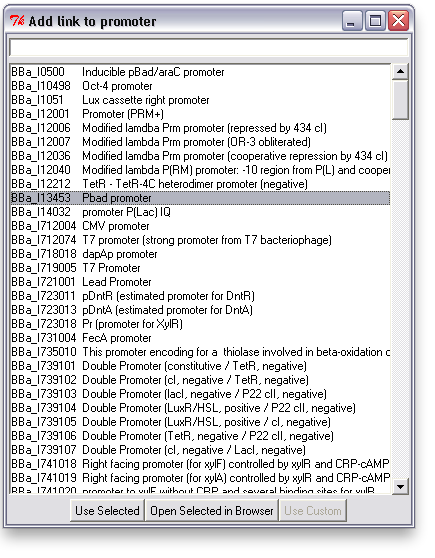
The first thing WikiDust does is allow you to associate TinkerCell items with parts on the registry. When the TinkerCell model is uploaded to a webpage, clicking on each item will go to the correct part page--in fact, many of the interactive diagrams on this wiki will be generated using WikiDust. The same linking mechanism could also be extended in the future to set the reaction kinetics, icon, etc. of the items. For now though, it's mostly a proof of concept demonstrating the use of a new semantic knowledgebase of parts.
Associating a link with an item is easy: just right click on an item and choose "Add Link". This will bring up a search window with suggested parts. You can search for a phrase, a BioBrick ID, etc. You can also open parts in a browser to read more about them or confirm that you've found the right one.
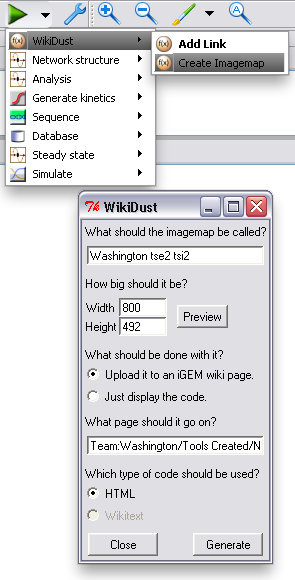
After the model of your device or system is finished, you can upload a representation of it to your page. WikiDust will automatically handle uploading to most [http://www.mediawiki.org/wiki/MediaWiki MediaWiki] sites, or it can just display code for you to copy and paste. By default, a download link for the complete model file is also included.
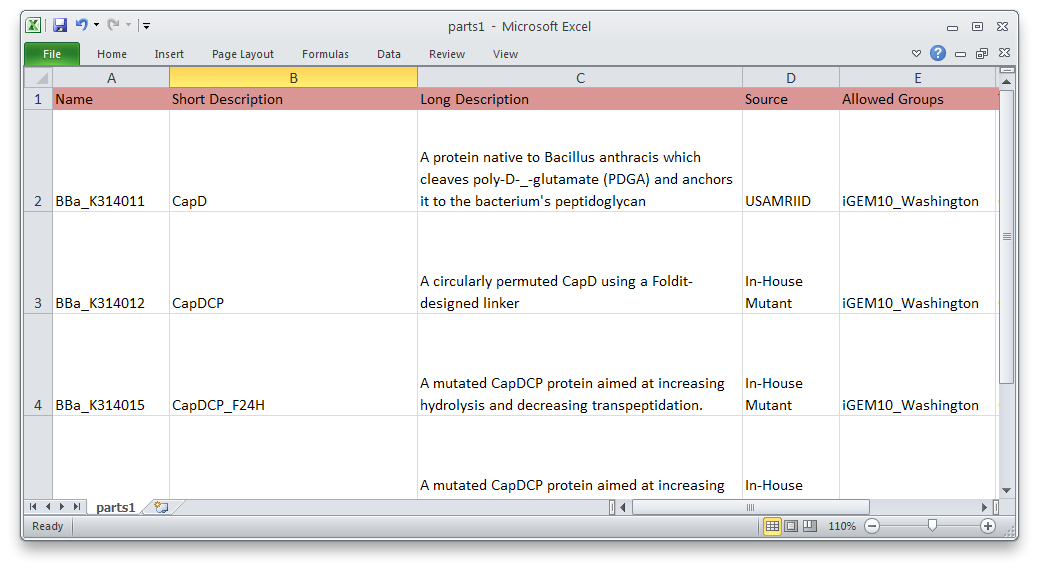
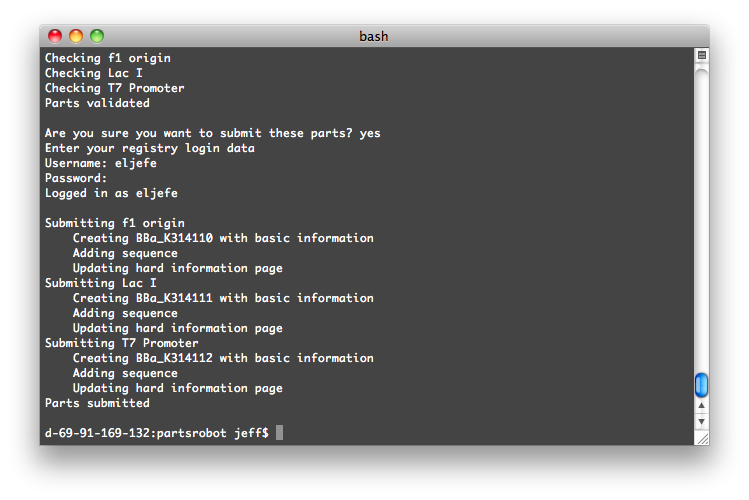
The UW team will also develop a program to submit multiple parts to the [http://partsregistry.org/Main_Page Parts Registry] at once. This project is in the early design phase now. It is expected to be useful for anyone who needs to submit a series of parts, for example if they are promoters of different strengths, variations on a protein, etc.
This will be a simple command-line tool. The user will make a spreadsheet with information about all the parts to be submitted. Then the tool will validate and submit each one to the registry.
 "
"