Switch
From 2010.igem.org
(→The switches) |
|||
| Line 6: | Line 6: | ||
=The switches= | =The switches= | ||
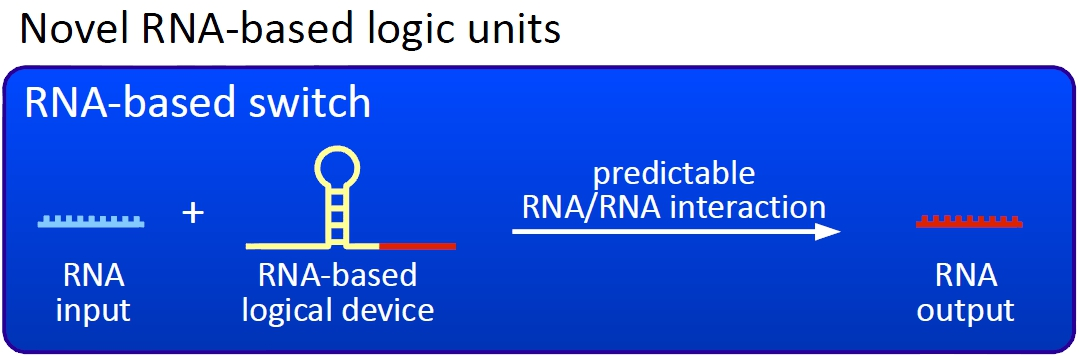
| - | Ou switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences, that are capable of stem loop formation, as transcriptional switches. | + | Ou switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences, that are capable of stem loop formation, as transcriptional switches.[[Image:TUM2010 novelunit.png|thumb|right|300 px|Novel switches based on RNA-RNA interaction]] |
| - | <br> | + | <br><br> |
We plan to control the termination at these switches with a small RNA molecule (our RNA "signal") that is complementary to a part of the stem loop forming sequence, inhibits the stem loop formation by complementary base-pairing and hence avoids transcription termination. The avaiablity of the complementary signal determines the way each switch operates. | We plan to control the termination at these switches with a small RNA molecule (our RNA "signal") that is complementary to a part of the stem loop forming sequence, inhibits the stem loop formation by complementary base-pairing and hence avoids transcription termination. The avaiablity of the complementary signal determines the way each switch operates. | ||
<br> | <br> | ||
| - | In a living cell, the production of a signal can be the response to a physical, chemical or biological stimulus that can be recognized by the cell via a receptor system. Several stimuli can induce the production of several | + | In a living cell, the production of a signal can be the response to a physical, chemical or biological stimulus that can be recognized by the cell via a receptor system. Several stimuli can induce the production of several signals, which can interfer with each other in a complex network with a final output depending on the initial stimuli. |
<br><br><br> | <br><br><br> | ||
Revision as of 15:31, 29 September 2010
|
||||||||
|
|
The switchesOu switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences, that are capable of stem loop formation, as transcriptional switches.
Note, that our systems work with termination on transcription level, so in our case the RNA-Polymerase falls off instead of stalling a ribosome. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the signal), which binds to the transcription mRNA and anticipates the formation of a stem loop. So basicly one gets a yes/no answer with a first switch: If the small RNA piece is available, transcription will continue. If not, it will be terminated. This is the way our first devices should work. Now, what's the great thing about our system? Well, in principle we can construct a lot of those switches. The only things you need are complementary RNA sequences and one of them must form a stem loop big enough to terminate transcription (switch) if the other one (signal) is not there. Those RNA-RNA-interactions can easily be calculated and RNA structures can be predicted with high accuracy - in complete opposite to anything based on proteins. So we are easily capable of scaling up our switch and add a couple more of those guys into a bacteria cell (they are really small, both switch and signal RNA). Since we can use RNA switches to regulate RNA output, we can easily built up networks, with our RNA pieces as the major controlling element. In comparison to proteins, which are normally used for work like this, RNA is predictable in its structure, fast in production and fast in degeneration, so quick and time-related responses are possible, non-toxic in all cases, really cute, available in all shapes, consists only of a few sugars, phosphates, and four bases, there is an nearly endless pool of possible switches... You need more? Well, one more we would really like to mention: RNA can be generated using DNA. Big news? It was in the Watson/Crick era! Well, since RNA can be generated using DNA, you can really really easily code for our RNA switches using certain DNA sequences. DNA is stable and quite easy to get into cells and wait, Biobricks consists of DNA! So this is how we would like to revolutionize Biobricks. We developed switches based on a very easy, yet totally new principle, which can be just cloned in between known Biobricks and offer totally new possibilities in combining bricks and gene regulation! Check here for more information, a cute Java applet to form your own network and enjoy!
|
|||||||
 "
"