Team:Tsinghua/Notebook/10 August 2010
From 2010.igem.org
(Difference between revisions)
(New page: == Module I, DT and Fan's part: == To ensure the result again, we decide to run a PCR with each gene’s own primers. PCR system (SuperMix): H2O 8.5μl primer1 0.5μl pri...) |
(→Module I, DT and Fan's part:) |
||
| Line 16: | Line 16: | ||
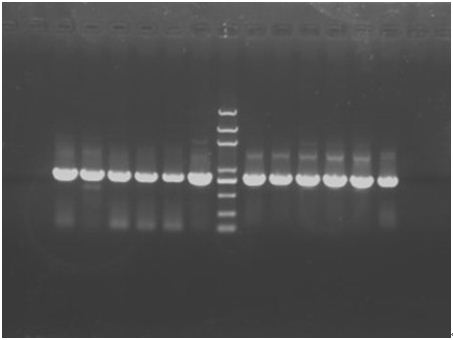
In the afternoon, when the PCR has finished, run a 1.2% gel. The result is shown below. | In the afternoon, when the PCR has finished, run a 1.2% gel. The result is shown below. | ||
| - | [[Image:10-8-10.jpg]] | + | [[Image:10-8-10.jpg]]<br/> |
Since the all 12 are all positive clones, stop the digestion and amplification step and go on the experiments. In consideration of the concentration of the plasmids and also the purity, we use P+E① and P+C③ undergo the follow steps. | Since the all 12 are all positive clones, stop the digestion and amplification step and go on the experiments. In consideration of the concentration of the plasmids and also the purity, we use P+E① and P+C③ undergo the follow steps. | ||
Revision as of 09:56, 3 September 2010
Module I, DT and Fan's part:
To ensure the result again, we decide to run a PCR with each gene’s own primers.
PCR system (SuperMix):
H2O 8.5μl primer1 0.5μl primer2 0.5μl template 0.5μl SuperMix 10μl Total 20μl
And just before receiving the result, we also go on the experiments repeat the preview steps in case that we get the all negative result. One is to select another 12 colonies to amplify; another is to digest the eGFP, chlr and pUC19 again with the same system shown before.
In the afternoon, when the PCR has finished, run a 1.2% gel. The result is shown below.
Since the all 12 are all positive clones, stop the digestion and amplification step and go on the experiments. In consideration of the concentration of the plasmids and also the purity, we use P+E① and P+C③ undergo the follow steps.
 "
"