Team:Tsinghua/Notebook/10 August 2010
From 2010.igem.org
Module I, group 2(b)
To ensure the result again, we decide to run a PCR with each gene’s own primers.
PCR system (SuperMix):
H2O 8.5μl primer1 0.5μl primer2 0.5μl template 0.5μl SuperMix 10μl Total 20μl
And just before receiving the result, we also go on the experiments repeat the preview steps in case that we get the all negative result. One is to select another 12 colonies to amplify; another is to digest the eGFP, chlr and pUC19 again with the same system shown before.
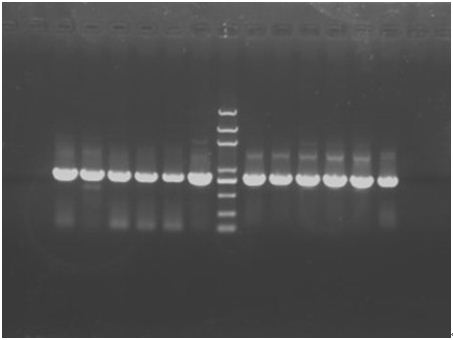
In the afternoon, when the PCR has finished, run a 1.2% gel. The result is shown below.
Since the all 12 are all positive clones, stop the digestion and amplification step and go on the experiments. In consideration of the concentration of the plasmids and also the purity, we use P+E① and P+C③ undergo the follow steps.
Double digest of the mCherry, P+E①, kan and P+C③ for each pair of restriction sites. Digest at 37℃.
Double digestion system:
mCherry:
H2O 6μl buffer BamHI 2μl mCherry 9μl SalI 2μl BamHI 1μl Total 20μl
P+E① for M:
H2O 11μl buffer BamHI 2μl P+E① 4μl SalI 2μl BamHI 1μl Total 20μl
kan:
H2O 0μl buffer BamHI 2μl kan 15μl KpnI 2μl BamHI 1μl Total 20μl
P+C③ for K:
H2O 11μl buffer BamHI 2μl P+C③ 4μl KpnI 2μl BamHI 1μl Total 20μl
Productions purify and measure the concentration. Ligate with Fermentas T4 ligase at 22℃ for 20min.
Ligation system:
P+E+M:
H2O 11.9μl 10×buffer 2μl M 1.6μl P+E for M 3.5μl T4 ligase 1μl Total 20μl
P+C+K:
H2O 14.4μl 10×buffer 2μl K 1.7μl P+C for K 0.9μl T4 ligase 1μl Total 20μl
Transform the ligation product into bacterial cells immediately. Spread about 100μl of the resulting solutions on LB plates (with 0.1% ampicillin).
 "
"