|
|
| Line 2: |
Line 2: |
| | Molecular tweezer tensile strength test. | | Molecular tweezer tensile strength test. |
| | | | |
| - | =Filamentous Cells= | + | =YneA= |
| - | ==Filamentous cells genes list==
| + | |
| - | | + | |
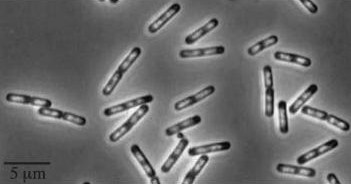
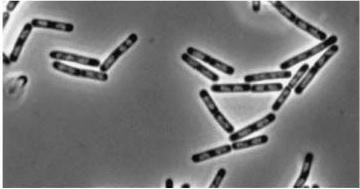
| - | *''yneA'' (Transcribed with ''yneB'', ''ynzC'', is an analogue of ''sulA'' in ''E.coli'') * our biobrick is designed to over express this gene reducing cell division possibly by inhibiting FtsZ ring formation or constriction.
| + | |
| - | *''dinR'' (Homologue of ''lexA'' in ''E.coli'' transcribed in the opposite direction)
| + | |
| - | *''ftsZ'' (Involved in the recruitment of other proteins to the divisisome for cytokinesis, strangely over expression results in disruption of Zring formation as well as reduced expression)
| + | |
| - | *''secA'' (Involved in the secretion of extracellular proteins and the insertion of transmembrane proteins)
| + | |
| - | * ''recA'' (Involved in SOS response removing the repressor DinR (LexA))
| + | |
| - | *''wpr'' and ''epr'' produce extracellular proteases that cleave the signal peptide/transmembrane domain of YneA
| + | |
| - | *''ezr'' produces a protein which sequesters FtsZ monomer by binding its C terminal domain and also inhibits GTP binding; however overexpression does not result in filamentation.
| + | |
| - | * ''min C,D ,J and divIVA'' prevent polar cell division .
| + | |
| - | *Positive regulators of FtsZ: ''ftsA, zapA, zipA, ftsL and divIC''
| + | |
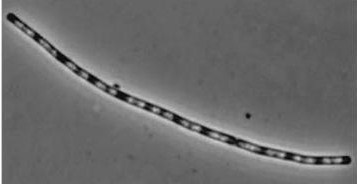
| - | *Inhibitors of Daughter cell separation: ''lytC,D,E,F and cwlS'' *Chains rather than filaments, ''yneA'' is also reported to increase the time spent in chains well into the stationary phase of bacterial growth.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
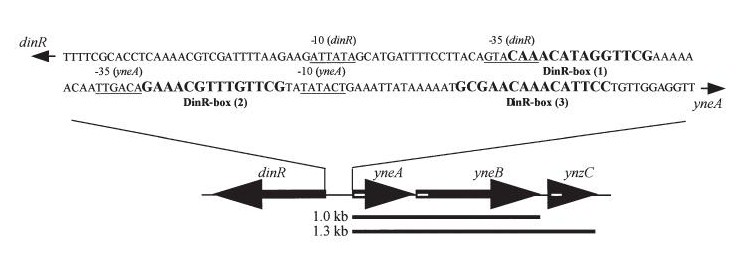
| - | ===YneA===
| + | |
| | {| | | {| |
| | |- | | |- |
| Line 68: |
Line 52: |
| | | | |
| | [[Media:yneA cloning strategy.pdf]] | | [[Media:yneA cloning strategy.pdf]] |
| - |
| |
| - | ===MinCDJ===
| |
| - |
| |
| - | {|
| |
| - | |-
| |
| - | |In ''B.subtilis'' cell division occurs precisely mid cell via the formation of the FtsZ ring (tubulin homologue), through Noc (Nucleoid occlusion: prevents division over nucleoids) and the min system (well described in E.coli), which prevents division taking place at the ends of the cell (poles).
| |
| - | |-
| |
| - | |Main function of Min system to prevent mini cell formation and ensuring only one cell division occurs per cell cycle.
| |
| - | |-
| |
| - | |Min’s role is not just in the inhibition of FtsZ. FtsZ recruits other components causing synthesis of the new wall and cell invagination.
| |
| - | |-
| |
| - | |[[Image:Min2.png]][[Image:Min.png]]
| |
| - | |-
| |
| - | |Cell division is regulated spatially and temporally.
| |
| - | |-
| |
| - | |Min C inhibits FtsZ ring formation; Min C interacts with Min D via its C-terminal domain. Min C inhibits lateral interaction between the filaments.
| |
| - | |-
| |
| - | | Min D is a membrane associated ATPase. (Min E ensures high concentrations of MinCD at the poles in ''E.coli'', no Min E homologue in ''B.subtilis!'' Instead DivIVA acts as a topological factor).
| |
| - | |-
| |
| - | |Min C in ''B.subtilis'' is responsible for localisation (shown using GFP) showed a primary site of localisation at the site of active division challenging the original model for the role of Min.
| |
| - | |-
| |
| - | |Min J was discovered linking Min D to DivIVA and therefore necessary for localisation.
| |
| - | |-
| |
| - | |Over expression of Min D in the absence of Min J causes lethal filamentation.
| |
| - | |-
| |
| - | |[[Image:TableMin.png]]
| |
| - | |-
| |
| - | |Min C sequence
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=2858550&to=2859298&strand=true&report=graph&content=5
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/gene/937500
| |
| - | |-
| |
| - | |MinD sequence
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=2857736&to=2858622&strand=true&report=graph&content=5
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/gene/937499
| |
| - | |-
| |
| - | |MinJ sequence
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=3620287&to=3621598&strand=true&report=graph&content=5
| |
| - | |-
| |
| - | |http://www.ncbi.nlm.nih.gov/gene/936668
| |
| - | |-
| |
| - | |'''van Baarle, S., & Bramkamp, M. (2010). The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PloS one, 5(3), e9850. doi: 10.1371/journal.pone.0009850.'''
| |
| - | |-
| |
| - | |-
| |
| - | |'''Bramkamp, M. & van Baarle, S., 2009. Division site selection in rod-shaped bacteria. Current opinion in microbiology, 12(6), 683-8. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19884039.'''
| |
Molecular tweezer tensile strength test.
YneA
Kawai, Y., Moriya, S., & Ogasawara, N. (2003). Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Molecular microbiology, 47(4), 1113-22. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12581363.
yneA cloning strategy
Media:yneA cloning strategy.pdf

 "
"