Team:Newcastle/16 July 2010
From 2010.igem.org
(Difference between revisions)
| Line 34: | Line 34: | ||
* '''Lane 9''': | * '''Lane 9''': | ||
* '''Lane 10''': | * '''Lane 10''': | ||
| - | * '''Lane 11'': | + | * '''Lane 11''': |
* '''Lane 12''': | * '''Lane 12''': | ||
* '''Lane 13''': | * '''Lane 13''': | ||
Revision as of 07:57, 9 August 2010

| |||||||||||||
| |||||||||||||
Contents |
LacI BioBrick Construction
Aims
- To use PCR to extract lacI (promoter, ribosome-binding site (RBS) & coding sequence (CDS)) from plasmid pMutin4 and ligate into vector pSB1AT3 in front of red fluorescent protein (RFP).
- The aim of today's experiment is to screen for the lacI insert. This could be done using PCR but that would require specific primers and is not as reliable.
Protocol
- Miniprep - the miniprep protocol was followed.
- Restriction digests - the restriction digests protocol was followed to allow the length of the DNA to be checked using single and double digest.
- Single digest with Pst1
- double digests with Pst1 and Xba1
- Gel electrophoresis - the protocol for gel electrophoresis was followed.
- Set up liquid broth culture in LB. The protocol for growing an overnight culture was followed.
Results
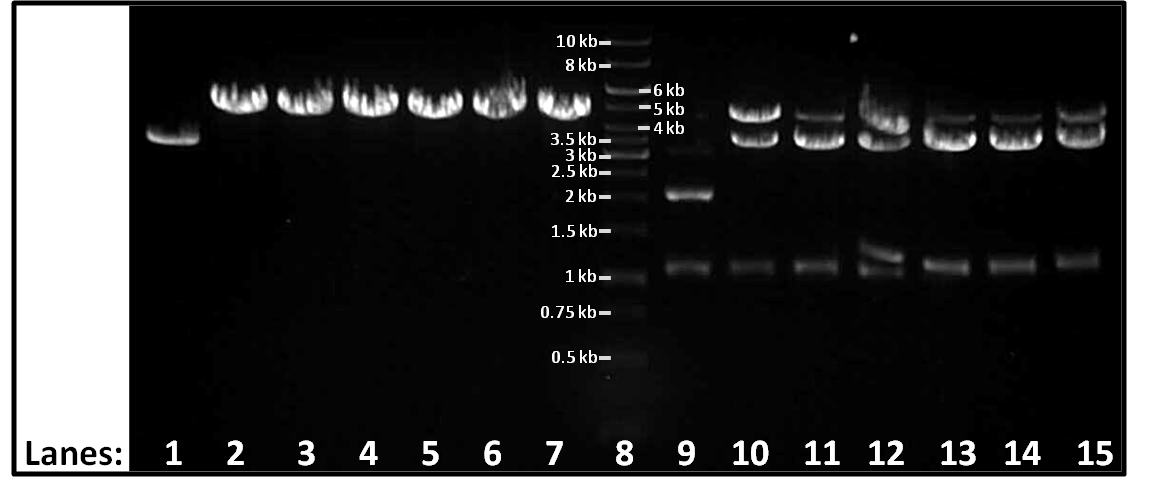
- The single digests (lanes 1-7) and the double digests (lanes (9-15) are separated by the molecular marker (lane 8).
Figure 1: Gel electrophoresis of the single and double digests using Pst1 and Xba1.
- Lane 1:
- Lane 2:
- Lane 3:
- Lane 4:
- Lane 5:
- Lane 6:
- Lane 7:
- Lane 8: 1kb DNA ladder
- Lane 9:
- Lane 10:
- Lane 11:
- Lane 12:
- Lane 13:
- Lane 14:
- Lane 15:
Inference
 
|
 "
"