Team:Newcastle/5 August 2010
From 2010.igem.org
Shethharsh08 (Talk | contribs) |
Shethharsh08 (Talk | contribs) |
||
| Line 54: | Line 54: | ||
=Amplification of Pspac_oid promoter by PCR= | =Amplification of Pspac_oid promoter by PCR= | ||
| + | |||
| + | ==Aim== | ||
| + | The aim of today's experiment is to amplify 6 different fragments for the construction of [[Team:Newcastle/Urease|''rocF'' BioBrick]] with the help of 6 different Phusion PCR. | ||
| + | |||
| + | ==Materials and Protocol== | ||
| + | Please refer to [[Team:Newcastle/PCR| PCR]] for Phusion PCR protocol. The details for the 6 PCR reactions are mentioned below: | ||
| + | |||
| + | ===PCR=== | ||
| + | {|border=1 | ||
| + | |- | ||
| + | !'''Tube''' | ||
| + | !'''Part to be amplified''' | ||
| + | !'''DNA fragment consisting the part''' | ||
| + | !'''Forward primer''' | ||
| + | !'''Reverse Primer''' | ||
| + | !'''Melting Temperature (Tm in °C) ''' | ||
| + | !'''Size of the fragment (in bp)''' | ||
| + | !'''Extension time* (in seconds)''' | ||
| + | |- | ||
| + | |1 | ||
| + | |Plasmid Vector | ||
| + | |pSB1C3 | ||
| + | |P1V1 forward | ||
| + | |P2V1 reverse | ||
| + | |58 | ||
| + | |2072 approx. | ||
| + | |60 | ||
| + | |- | ||
| + | |2 | ||
| + | |Pspacoid Promoter | ||
| + | |pMutin4 | ||
| + | |P1P1 forward | ||
| + | |P2P1 reverse | ||
| + | |49 | ||
| + | |106 approx. | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |1st fragment of ''rocF'' CDS | ||
| + | |''B. subtilis'' 168 chromosome | ||
| + | |P1S1 forward | ||
| + | |P2S1 reverse | ||
| + | |58 | ||
| + | |246 approx. | ||
| + | |15 | ||
| + | |- | ||
| + | |4 | ||
| + | |2nd fragment of ''rocF'' CDS | ||
| + | |''B. subtilis'' 168 chromosome | ||
| + | |P3S2 forward | ||
| + | |P4S2 reverse | ||
| + | |65 | ||
| + | |597 approx. | ||
| + | |20 | ||
| + | |- | ||
| + | |5 | ||
| + | |3rd fragment of ''rocF'' CDS | ||
| + | |''B. subtilis'' 168 chromosome | ||
| + | |P5S3 forward | ||
| + | |P6S3 reverse | ||
| + | |66 | ||
| + | |125 approx. | ||
| + | |15 | ||
| + | |- | ||
| + | |6 | ||
| + | |Double Terminator | ||
| + | |pSB1AK3 consisting BBa_B0014 biobrick | ||
| + | |P1T1 forward | ||
| + | |P2T1 reverse | ||
| + | |56 | ||
| + | |116 approx. | ||
| + | |15 | ||
| + | |} | ||
| + | |||
| + | '''Table 1''': Table represents 6 different Phusion PCR reactions and the parts which are amplified so that they can be ligated together with the help of Gibson Cloning method. | ||
| + | * The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different. | ||
| + | * For learning about the ''rocF'' fragments, please refer to the [[Media:Cloning_strategy_for_rocF.pdf|Cloning strategy for ''rocF'']]. | ||
| + | |||
| + | ==Discussion== | ||
| + | All the 6 Phusion PCR reactions were done however, gel electrophoresis was not done today to check whether the fragments have actually amplified or not. | ||
| + | |||
| + | ==Conclusion== | ||
| + | Tomorrow 5th August, 2010, we would be running gel electrophoresis to check the outcome of the 6 PCR reactions and later all the fragments will be ligated with help of Gibson protocol. | ||
| + | |||
Revision as of 00:37, 6 August 2010

| |||||||||||||
| |||||||||||||
Contents |
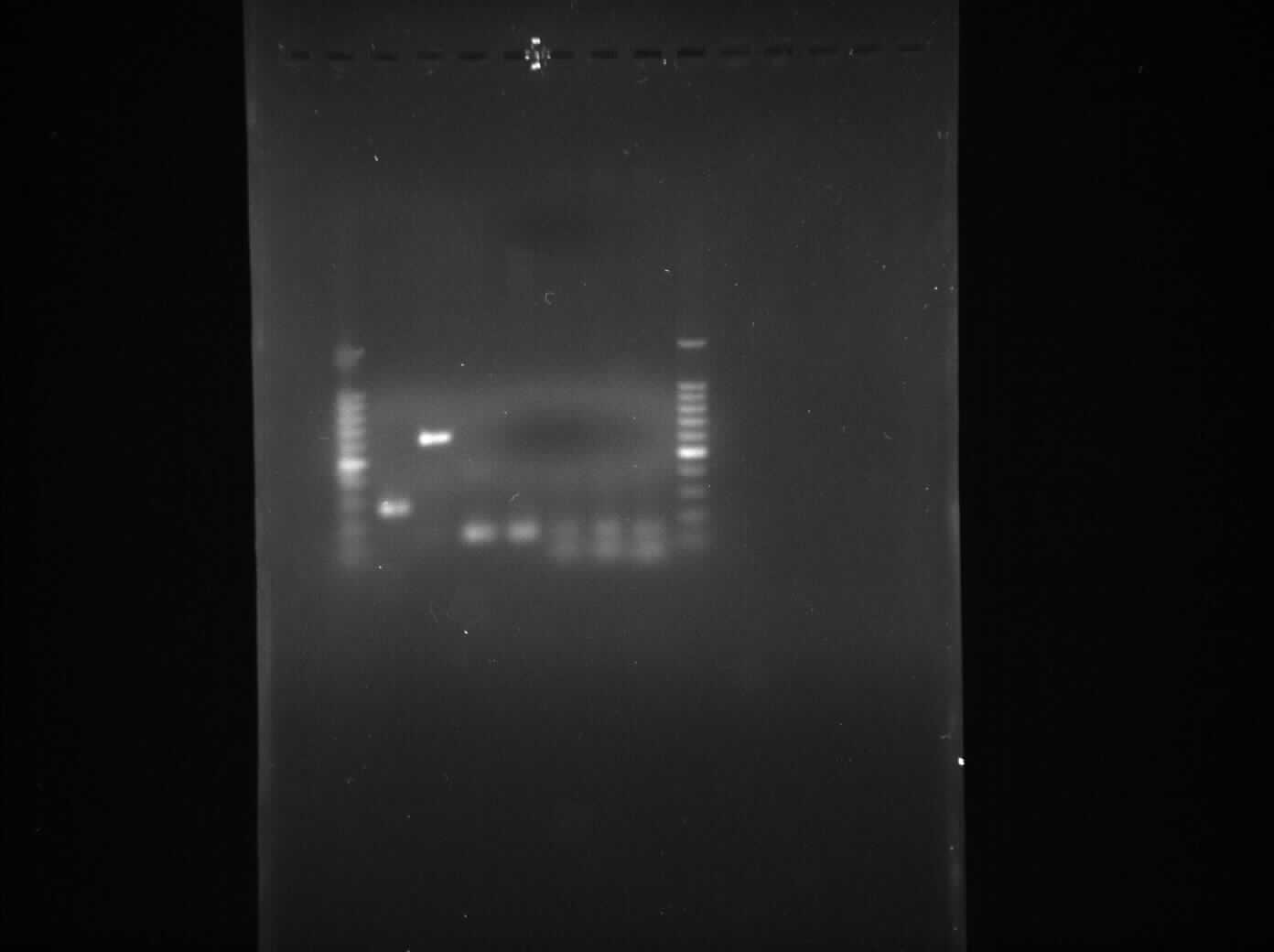
Gel Electrophoresis for the Amplified Fragments of rocF
Aim
The aim of the experiment is to perform gel electrophoresis for all the 6 PCR reactions which took place yesterday 4th August, 2010 and thus confirm that all 6 PCR reactions were successful.
Materials and Protocol
Please refer to: Gel electrophoresis.
Result
- Lane 1: 1kb DNA ladder
- Lane 2: BioBrick compatible vector pSB1C3
- Lane 3: Pspac_oid promoter
- Lane 4: 1st fragment of rocF CDS
- Lane 5: 2nd fragment of rocF CDS
- Lane 6: 3rd fragment of rocF CDS
- Lane 7: Double Terminator
- Lane 8: 1kb DNA ladder
| Biobrick compatible vector pSB1C3 | Pspac_oid pormoter | 1st fragment of rocF CDS | 2nd fragment of rocF CDS) | 3rd fragment of rocF CDS | Double Terminator | |
|---|---|---|---|---|---|---|
| Size of the Fragment (in bp) | 2072 approx. | 106 approx. | 246 approx. | 597 approx. | 125 approx. | 116 approx. |
Table 1: Table represents the size of the fragments represented as bands on the gel in their corresponding lanes.
Discussion
We found bands in the lanes 2,4,5,6 and 7 of the correct sizes but lane 3 did not contain any band.
Conclusion
This experiment shows that the PCR reaction was successful for all the fragments apart from Pspac_oid promoter which was present in the lane 3 and did not show any band. This could be because of the following problems:
- Primer sequences could be incorrect.
- Melting temperature could be incorrect.
- Plasmid pMutin4 could have degenerated due to long term storage.
Solution for the problem
- If P1 buffer of the Qiagen miniprep kit is contaminated, then use a different kit. We have Promega miniprep kit which will be used tomorrow.
- If RNAse enzyme is inactive, then add extra RNAse into the P1 buffer. We would be adding 10 µl in the P1 buffer solution.
Amplification of Pspac_oid promoter by PCR
Aim
The aim of today's experiment is to amplify 6 different fragments for the construction of rocF BioBrick with the help of 6 different Phusion PCR.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol. The details for the 6 PCR reactions are mentioned below:
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | pSB1C3 | P1V1 forward | P2V1 reverse | 58 | 2072 approx. | 60 |
| 2 | Pspacoid Promoter | pMutin4 | P1P1 forward | P2P1 reverse | 49 | 106 approx. | 15 |
| 3 | 1st fragment of rocF CDS | B. subtilis 168 chromosome | P1S1 forward | P2S1 reverse | 58 | 246 approx. | 15 |
| 4 | 2nd fragment of rocF CDS | B. subtilis 168 chromosome | P3S2 forward | P4S2 reverse | 65 | 597 approx. | 20 |
| 5 | 3rd fragment of rocF CDS | B. subtilis 168 chromosome | P5S3 forward | P6S3 reverse | 66 | 125 approx. | 15 |
| 6 | Double Terminator | pSB1AK3 consisting BBa_B0014 biobrick | P1T1 forward | P2T1 reverse | 56 | 116 approx. | 15 |
Table 1: Table represents 6 different Phusion PCR reactions and the parts which are amplified so that they can be ligated together with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different.
- For learning about the rocF fragments, please refer to the Cloning strategy for rocF.
Discussion
All the 6 Phusion PCR reactions were done however, gel electrophoresis was not done today to check whether the fragments have actually amplified or not.
Conclusion
Tomorrow 5th August, 2010, we would be running gel electrophoresis to check the outcome of the 6 PCR reactions and later all the fragments will be ligated with help of Gibson protocol.
Concrete Tensile Splitting Test
Aim
To get samples of cracked concrete for Bacilla Filla to fill up the cracks and to determine the tensile strength of concrete before and after the cracks are filled up.
Materials
- Concrete cylinder
- Jubilee clips
Procedure
 
|
 "
"