Team:Calgary/22 July 2010
From 2010.igem.org
Raidakhwaja (Talk | contribs) |
Raidakhwaja (Talk | contribs) |
||
| Line 14: | Line 14: | ||
<u>Chris</u> | <u>Chris</u> | ||
| - | Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C | + | Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C. |
<u> Raida </u> | <u> Raida </u> | ||
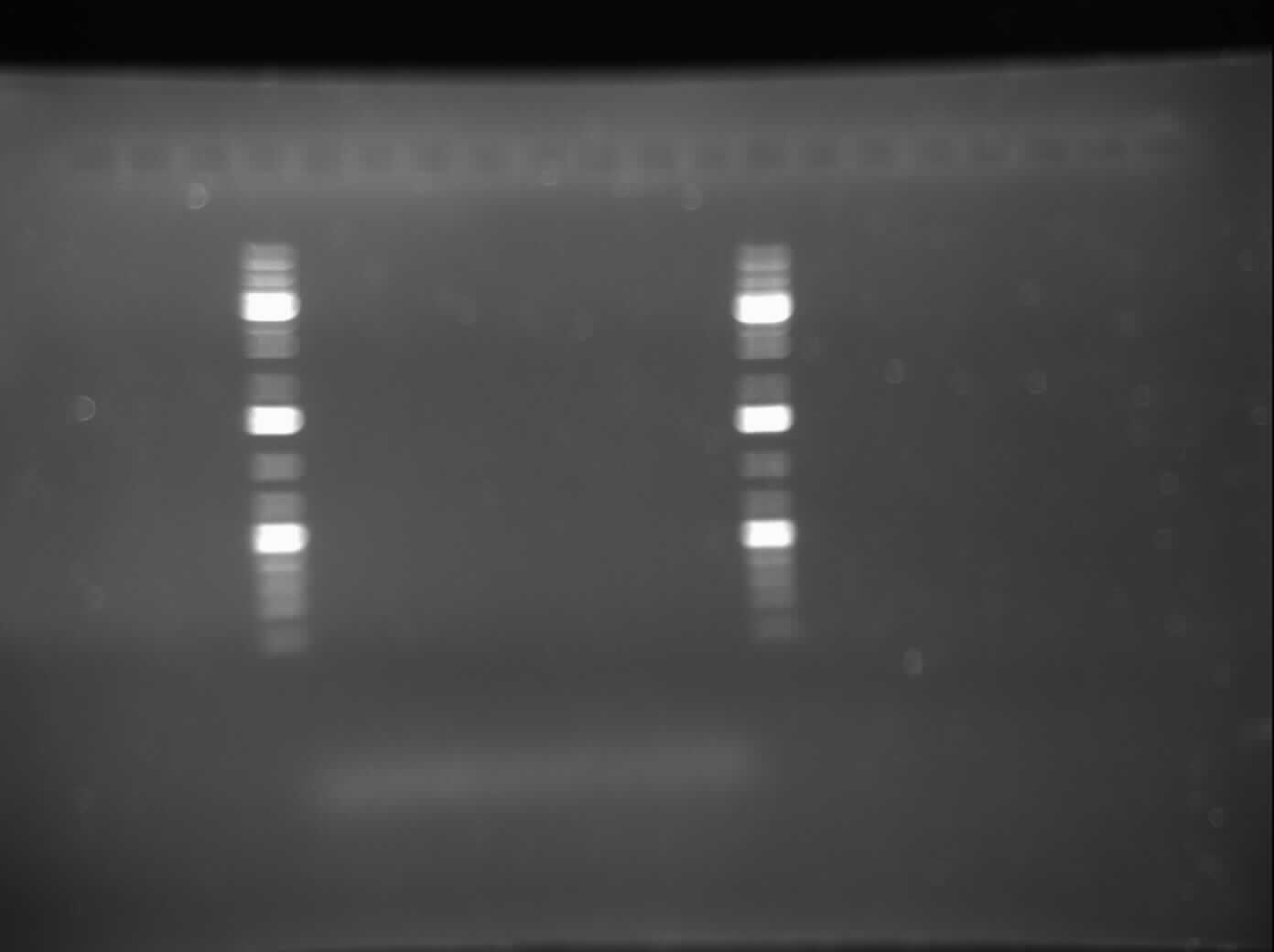
| - | Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. | + | *Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. I am also going to change the buffer because the picture came out blurry and I am predicting that it is because of the buffer. |
Please refer to the image of the gel to the side. | Please refer to the image of the gel to the side. | ||
| + | |||
| + | * Near the end of the day, with the help of my supervisor, Henry, I set up and ran another PCR with the same DNA and the same primers. This time, the difference was that I used the Qiagen Master Mix. I was also suggested to change the annealing temperature to 52 degrees for 30 seconds instead of 55 for 1 min as I on June 21st. I will run a gel electrophoresis of my PCR product tomorrow to check whether the MalE gene specific primers annealed at the right location and amplified Mal-E gene. | ||
| + | |||
}} | }} | ||
Revision as of 19:18, 23 July 2010

Thursday July 22, 2010
Himika
Today I miniprepped 10 colonies from the I0500-B0034 construction using Sigma-Aldrich kit. I also did a plasmid switch of I0500-B0034 from pSB1A3 to pSB1AC3.
Jeremy
Today I did a colony PCR of K23900+I13507, K239000+I13504, K135000+I13507, K135000+I13504 and prepared an overnight of several colonies. I also re-ligated and transformed I0500+B0034 in pSB1AC3 plasmid.
Chris
Today, I calculated the values of the enzymes we received from New England Biolabs. As well, I redid the gel of the colony PCR of the CpxP promoter with a 1.0% gel. This time, the DNA actually moved and the picture of the gel can be seen on the right. However, the bands were very non-specific. To optimize, I set up one gradient PCR with temperature of annealing step ranging from 53-58°C.
Raida
- Today, I did a gel electrophoresis of my PCR product from June 21, which was done to test the MalE primers. The results were negative as no bands were seen. This indicated that there was no amplication. I will do another PCR with the annealing temperature changed. I am also going to change the buffer because the picture came out blurry and I am predicting that it is because of the buffer.
Please refer to the image of the gel to the side.
- Near the end of the day, with the help of my supervisor, Henry, I set up and ran another PCR with the same DNA and the same primers. This time, the difference was that I used the Qiagen Master Mix. I was also suggested to change the annealing temperature to 52 degrees for 30 seconds instead of 55 for 1 min as I on June 21st. I will run a gel electrophoresis of my PCR product tomorrow to check whether the MalE gene specific primers annealed at the right location and amplified Mal-E gene.
 "
"