Lab notes (7/12 - 7/18)
Group: Flagella
Exp. 1
Methods: [MiniPrep], NanoDrop and gel electrophoresis
Notes: We used 9 ml ON culture (Lag phase cells), loaded 2ul sample and 8ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker
Results: Nanodrop: Tube 1: 30.7 mg/ul Tube 2: 27.4 mg/ul
Gel electrophoresis: No result
--Louch07 15:00, 9 July 2010 (UTC)
Exp. 2
Methods: gel electrophoresis
Notes: We ran another gel electrophoresis on the miniPrep sample form above. But now we loaded 4ul sample and 4ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker.
Results: Gel electrophoresis: Bands were detected. The psb3k3 plasmid is 2750bp long and the RFP with generator is 1096 bp, which gives a band at about 4000bp compaired with the marker.
Exp. 3
Methods: [MiniPrep], NanoDrop and gel electrophoresis.
Notes: We used 10 ml af a culture in Log phase (1ml cells from an ON culture + 9ml LB medium, incubated at 37 degrees celcius for 4 hours), loaded 4ul sample and 4ul agarose loading dye on a 1,5% gel,used a DNA ladder mix (100-10000 nucleotides) as marker.
Results: Nanodrop: Tube 1: 38.7 mg/ul Tube 2: 32.4 mg/ul.
Gel electrophoresis: Bands were detected.
--Louch07 17:21, 12 July 2010 (UTC)
Polyferation of FlhDC, FlhD and FlhC genes
Methods: [PCR] and Gel electrophoresis.
Notes: We decided to test our primers on previously purified cromosomal DNA. Examination of the primers showed that the FlhC reverse primer had a melting temperature of only 45˚C. Therefore we decided to run the samples on a gradient PCR. Simultaneously, we prepared 2 extra samples, isolating FlhD and FlhC, respectively. We did this because we wanted to see if our problems were caused because the combined gene-sequence was to long (932bp).
Because we just wanted to test our primers in this PCR, we used Taq polymerase, because although it doesn’t proofread, it is remarkably cheaper than Pfu. On the [http://www.fermentas.com/en/products/all/pcr-qpcr-rt-pcr/standard-pcr/ep028-taq-dna-native Fermentas homepage] we found that the annealing temperature for Taq is Tm-5 , which in this case means 40˚C. However, Taq polymerase is not very effective at temperatures under 50˚C so we designed the gradient to lies between 40 and 55˚C. The PCR machine chose the following temperatures:
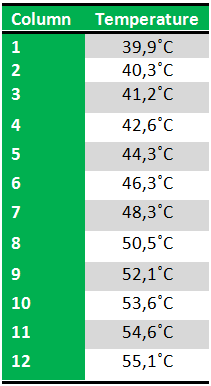
We then chose six temperatures to test:
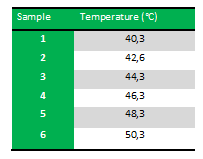
The following table shows the Gradient PCR program:
Results: The experiment was succesfull! We could detect FlhDC DNA at temperatures between 42.6˚C and 48.3˚C. FlhD DNA at temperatures between 40.3˚C and 44.3˚C and also between 48.3˚C and 50.3˚C. FlhC DNA at temperatures run between 40.3˚C and 50.3˚C.
--Louch07 10:13, 12 July 2010 (UTC)
Polyferation of FlhDC with Pfu
Date: The 12th of july
methods: colony PCR and gel electophoresis
Notes: The purified chromosomal DNA was tested with Pfu; a polymerase with proofreading ability. The DNA was from the E. coil strain MG 1655 (tube 17) The annealing temperature was used fore the PCR was 44˚C. We decide use this annealing temperature while it is in the middle of the temperature span we successful used in the last experiment. Further we increas the elongation time to 1 min and 30 sec. Other than these changes we followed the PRC program in the protocol. the PCR product was run on a 1,5% agarose gel.
results: Unfortunately the experiment did not give any results on the gel.
--Pernm07 10:56, 13 July 2010 (UTC)
Polyferation of FlhDC with pfx
Date: The 13th of july
methods: The experiment was perform in the E.coli strain, MG1655. The DNA purification was done in duplicates and are saved in the freezer in tube 8 and 9. (green label). There DNA concentration was measure by Nano Drop and there contantretion are 16,87ng/uL and 19,25ng/uL, respectively.
protocol: Genomic DNA purification and colony PCR.
Notes: The experiment was generally preformed corresponding to the protocol but small modification was made. In the DNA purification experiment we used 300uL instead of 200uL of the overnight culture and it is important to remember that the 800uL precipitation sulution, containing 80uL solution and 720uL deionized water, are made directly before use.
because our PRC for the time being havent been successful we are trying to do PCR with pfx wich are another polymerase with proofreading ability. The PRC program used for running PRC with pfx are as described below:
1
|
start
|
94°C
|
2 min
|
2
|
Denaturing
|
94°C |
1min
|
3
|
Annealing
|
48°C |
1 min
|
4
|
Elongation
|
68°C |
2 min
|
5
|
GOTO 2
|
|
Rep x 29
|
6
|
End
|
68°C |
3min
|
7
|
Hold
|
4°C |
|
results: there did not appear any lanes when we runed the gel electophoresis. Therefore the next step is to run PCR with Taq polymerase to check if the problem are the pfu polymerase or if the DNA is not purify properly.
--Pernm07 10:19, 13 July 2010 (UTC)
Group: Photosensor
Group: Retinal
Colony PCR of K274210 in pSB1A2 (transformation from 08/07)
Start date: 12/07 End date: 12/07
Methods: [Colony PCR]
Protocol: Modified iGEM colony PCR protocol
Notes: MgCl2 was added as a gradient,sample 1-2 contain 2 ul (0,04255), sample 3-5 2,25ul (0,04787). We mistakenly added 30ul (instead of 17ul) premix to sample 6 - 9, so the concentration of MgCl2 is reduced. Sample 6 contains 2,25ul (0,0375), Sample 7-8 2,5ul (0,04167), Sample 9 2,75ul (0,04583).
Gel was loaded with DNA-ladder plus, with the upper marker at 5000bp.
Results:
We found plasmids at the expected 5kbp marker in colonies 1-8.
Transformation of BBa_R0011 and BBa_I0500
Start date: 12/07 End date:
Methods: ON culture; [Competent cells of E.Coli for transformation]; [Transformation of E.Coli].
Protocols: iGEM protocol for competent cells, iGEM protocol for transformation of E.Coli.
Experiment done by: Christian, Maria and LC (Making competent E.Coli)
Date: 12/07
Notes: Everything was done after protocol.
Results:
Analysis:
Experiment done by: Christian, Maria and LC (Transformation of pBad and lacl promoter)
Date: 12/07
Notes: Added 200ul of the un-pelleted cells, instead of 150ul to the agar plates.
Some of the agar dishes were damaged during drying and plating. We tried to use them anyways.
Results:
Analysis:
Mini-prep of pSB1A2 w. B0034, pSB1AK3 w. B0015 and pSB3K3 w. J04450(transformation from 08/07)
Start date: 12/07 End date: 12/07
Methods: [MiniPrep]
Protocol: Fermentas protocol
Notes: pellet from 10 mL ON-culture was resuspended in 500uL resuspension buffer, and transferred into two eppendorf tubes, which were run in parellel
samples were loaded into a 1% gel with a gene ruler red marker.
Results:
Transformation of MG1655 e. coli with BBa_274210, BBa_E0040 and BBa_I0500
Start Date: 13/07 End Date:
Methods: ON cultures, Competent Cells, Transformation of competent cells.
Protocols:CC1.1, TR1.1
Experiment: Making Competent e. coli MG1655
Date: 13/07
*ON cultures of strain MG1655 were put over the night before.
Notes: Eksponential growth was started at 09.30 with an OD of 0.19A. The cells were harvested at 10.26. OD was .048 at 10x dillution.
Cells were put in fridge at 11.00. They will be ready at 11.30 and good for 48 hours.
Results: Competent cells were made according to protocol. Apparently MG1655 strain reaches OD 0.5 at least an hour faster than TOP 10.
Analysis:It is important to measurement at 10 minute intervals after OD 0.20 when growing MG1655 strain.
Experiment: Transforming MG1655 strain with BBa_E004 (GFP coding sequence) and BBa_I0500 (pBad arabinose inducible promoter)
Date: 13/07-2010
Notes:
I0500 is located in pSB2K3. E0040 is in pSB1A2. 200ul cells were added to each tube. Positive control is RFP generator as usual. Negative control is just cells.
Only 1ul DNA material was transferred per tube, since it was taken from distribution plates.
Plates were dried at 42°C for 15 minutes, just prior to plating. Upon plating we discovered a problem with many of the ampicilin plates we made a couple of days ago. The agar breaks on the slightest contact, and are therfore impossible to use.
As a result of the defective agar plates, most of our cells carrying BBa_E0040 were ruined. We managed to save and plate out two batches, one from each tube, so we are hoping for the best tomorrow.
Miniprep of BBa_K098995 in pSB1A2
Start date: 13/07 End date: 13/07
Methods: Fermentas GeneJET plasmid miniprep kit
Protocol: MP1.1
Experiment done by:"" LC
Notes: Centrifuged the ON for 15 mins. at 3000 rpm. Loaded 10 ml of culture in each tube and added 500ul resuspension solution, then split it into two samples.
Results:
Results were as expected (correct), the plasmid was around 3000 bp long on the gel.(Insert Gel picture and nanodrop results table)
Analysis:All four samples turned out fine, so they were pooled and frozen. (for details see results)
Nr. 28 in the freezer = BBa_K098995
 "
"


