Team:Edinburgh/Notebook/BRIDGE
From 2010.igem.org
| Line 43: | Line 43: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Genomic">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Genomic">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Project/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 50: | Line 50: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial" class="dir">bacterial BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Core_repressilator">the project</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_producer">red light</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Red_light_sensor">red sensor</a></li> | ||
| Line 59: | Line 59: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/BioBricks#Bacterial">submitted parts</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Bacterial">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Bacterial/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 72: | Line 72: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Tools">tools</a></li> | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Modelling">results</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">future | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/Future">the future</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Modelling/References">references</a></li> | ||
</ul> | </ul> | ||
| Line 79: | Line 79: | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human" class="dir">human BRIDGEs</a> | ||
<ul> | <ul> | ||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Communication">communication of science</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Branding">iGEM survey</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Conversations">conversations</a></li> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/Epic">the epic</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/ | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/FutureApps">future applications</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Edinburgh/Human | + | <li><a href="https://2010.igem.org/Team:Edinburgh/Results#Human">further thoughts</a></li> |
<li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | <li><a href="https://2010.igem.org/Team:Edinburgh/Human/References">references</a></li> | ||
</ul> | </ul> | ||
Latest revision as of 02:11, 28 October 2010
BRIDGE
5/7/2010
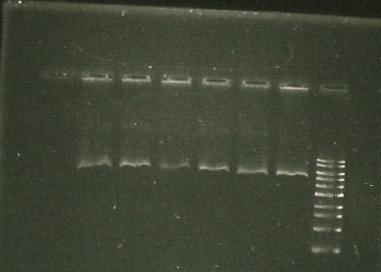
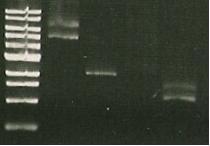
- sacB digest with EcoRI -band around kb (gel 1)
6/7/2010
- sacB digest with EcoRi/PstI. Band also around 5 kb.
7/7/2010
- PCR of sacB to check insert. Used sacB tube 1 and 3.
- Restriction digest of PCR with EcoRI and NotI.
- lane 1- marker
- lane 2, 3, 4 - sacB 1, 2 & 3 cut with NotI; band size: slightly bigger than 4 kb
- Lane 5- PCR of LuxAB; band size around 1.5. kb
- lane 6- Vector of sacB (pSB1C3, 2072bp, aroun 3kb with RFP) cut with NotI; band size around 1.5. kb
- Lane 7- PCR of sacB with gene sepcific primers; band size around 1.5. kb
- Trouble shooting: sacB cut with NotI give about 4kb, whereas cutting with EcoRI gives 5.5. kb... hmhmhm.
there should be 2 NotI sites on either side of the insert but we only get 1 band--> is there only 1 NotI site? Furthermore, PstI/EcoRI digests give only 1 band (same as cut only with EcoRI), so maybe there is no PstI/NotI site?
- Purifying sacB from PCR product to start making BRIDGE- row F in the BOX 1.
9/7/2010
- Making working solutions of primers from stock: 4ul of H20; 1 ul of stock solution
- Primers: forward 72.3 C, revers- 77.6 C; length of the product: 2.6-2.7 kb
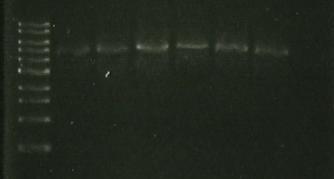
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 | Lane8 |
| ladder | sacB | sacB digest wtih EcoRI-HF | sacB + | digest `5009 | S248T PCR product | Pure 356K | Pure 356R |
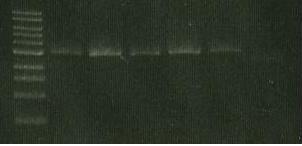
- redo sacB- catR ligation
- Run 5 ul of ligation on gel to check for large bands
- redo RLS part transformations
12/7/2010
- Transformants of luxAB-edi1 failed- probably smth wrong at the purification stage...
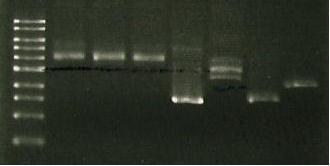
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 |
| ladder | luxAB-ediI digest | luxAB-ediI ligation | sacB digest | catR digest | sacB- catR lgation |
| expec. no of bands | 2 | 1 or 3 | 1 | 1 | 1 band= to 4+5 |
- If all failed- purification procedure failed
- If 3 and 6 failed- ligation failed
- If all fine then PCR/transformation failed
conclusion:...
- 3 and 6 failed- ligation problem (not enough DNA, ligase).
- Band 1- luxAB but no ediI (?)
- Repeated digest (sacB with XbaI, catR with SpeI, LuxAB & EdiI with SpeI & SacI) and purification.
- Set up ligations with 2ul ligase instead of 1 ul (1ul of H2O less). If this fails- perhaps only 2-3 h ligation ina RT (SpeI is set up in both digest- could be common cause)
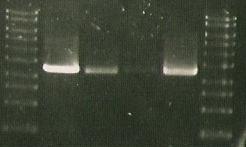
13/7/2010
- First, run PCR of catR-sacB ligation (use DNA?????' MK1- with sacBr1 (19.6.10); MK2- sacBr1(10)
- Gel electrophoresis of: digested catR, digested sacB, ligated sacb-catR, digested LuxAB-ediI and ligated luxAB-ediI along with sacB-catR PCR
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 |
| ladder | ediI | digested LuxAB-ediI | ligated luxAB-ediI | ligated catR & sacB | PCR 1 (MK1) | PCR 2 (MK2) |
'AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAARGH!!!!!!!!!!!!!!!1'
- Almost completely convinced it's the ligase... or me (MK). Officially taking myself off the project. xx
- (Spotted 2 purified sacBs in the freezer... maybe try the original if this one fails...?)
3/8/2010
- minipreps: marker (lane 1& 8) & sacb 8.1-8.6( Lanes 2-7)
Gel with :
- cat + sacB (PCR product) (6)
- vector + cat + sacB (7) (both done by CF)
20/8/2010
- Made plates variety of sucrose combinations--> 40 cml
- 0%
- 0.1%
- 0.25%
- 0.5%
- 1.0%
- Plated E6 miniprep, which should show some sucrose sensitivity (1plate at each concentration)
- Control- E5, 1 plate at each concentration
30/8/2010
- Primers have arrived for up and downstream sequences of TnaA region.
8/9/2010
- Lane 1 &8- marker, Lane 6- cat +sacB PCR; Lane 7- vector+cat+sacB
13/9/2010 Plane for week begininng 13th:
- Amplify RFP with primers --> RBS-F2; YFP - R2 ---> to amplify RFP
- Digest UP with SpeI and B-D with XbaI
- Ligate UP to BRIDGE-DOWN
- Digest RFP with XbaI and SpeI
- Ligate UP and RFP
- Ligate DOWN and RFP
- Also- registry editing for RLS (definately dodgy sequence), LovTap
- Justification of reseubmission
- Look up absorption/emission rates and transmission through medium for Donal
20/9/2010
- Achieved: cat/sacB- traA down ligation
- Attempted: traA up- cat/sacB/down ligation; traA up- RFP ligation; RFP- traA down ligation/ ALL FAILED
- Running: purified digest of:
- cat/sacB + SpeI
- tnaA down- XbaI
- tnaA up + SpeI
- cat/sacB/down + XbaI
- RFP + XbaI
and also running: UP- BRIDGE- DOWN ligation; UP - RFP ligation
Have: PCR of cat/sacB, RFP, BRIDGE-DOWN, traA up, traA down; ligation of BRIDGE-DOWN
Run gel:
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 | Lane 8 |
| Ladder | Pure SacB | Pure SacB & EcoRI | sacB & catR PCR | I15009 digest | S284T PCR Product | Purified 356K | Purified 356R |
25/10/10
Repetitive failure of purifications with both traditional method and kits, unsure what is going wrong. Have decided to tell Chris. Off to Firbush on Wednesday.
01/10/10
In my absence (at Firbush with the rest of the Biotechnologists) Chris has managed to construct the tnaA UP-cat-sacB-tnaA DOWN segment that we need for the first part of the BRIDGE protocol. I will continue to build the tnaA UP-RFP-tnaA DOWN segment.
Set up ligations of RFP-DOWN and UP-RFP, left in 16C incubator overnight.
02/10/10
Lab on a Saturday again, fun fun fun...
Retrieved and amplified ligations and ran on gel. UP-RFP appears to be significantly shorter than RFP-DOWN and indeed shorter than it ought to be (should be about 2kb, is only around 1.5kb).
Purified both products, then digested RFP-DOWN with XbaI. (Used 2hour incubation time to write presentation for Wednesday).
Set up ligation of UP with RFP-DOWN and left in incubator overnight.
03/10/10
Bus to Darwin for 11:20am; enter Darwin; lift to 8th floor; retrieve ligations from waterbath in cold room; walk down to 7th floor; put ligation in freezer; lift to ground floor; leave Darwin; catch 11:40am bus back to central.
04/10/10
PCR of ligation of UP-RFP-DOWN. Made and ran gel along with purified PCRs from Saturday, just in case they're still not working.
They worked! The purifications were perfect. There is a band about the size of UP-RFP-DOWN in it's lane, but it's quite faint. I will probably repeat the PCR with a lower annealing temperature (around 50) this afternoon. I suspect the UP primers do not anneal properly above 54C.
07/10/10
Chris set up overnight cultures of DH5-alpha cells and JM109 cells, both containing the lambda-red plasmid, in 2.5ml LB with tetracycline.
Repeated PCR of up-RFP-down ligation as well as PCR of 3 subcultures suspected to contain the cat-sacB construct, performed using the tnaA UP forward primer and the tnaA DOWN reverse primer.
08/10/10
Took 2 5ml LB bottles:
- Added 1.7 microlitres of tet15 and 0.3ml DH5-alpha overnight culture to one
- Added 5 microlitres of tet15 and 0.3ml of JM109 overnight culture to the other
Grew second cultures for 2 hours at 30C with shaking
Measured optical density after 2 hours:
- Water = 0.00
- DH5-alpha = 0.251
- JM109 = 0.320
Added 100microlitres arabinose to both cultures and grew at 37C for one and a half hours with shaking.
Prepared cells for electroporation:
1- Added 1.5ml of liquid culture to an eppendorf
2- Spun down cells at 11,000rpm for 30s
3- Removed supernatant, added 1ml of cold sterile water and resuspended cells
4- Repeated steps 2 and 3 twice more
5- Added 40microlitres of suspended cells and 1microlitre of DNA construct (up-cat-sacB-down) to chilled electroporation cuvette (also prepared one cuvette of each cell culture without DNA for controls)
Performed electroporation
Suspended cells in cuvette by adding 0.5ml LB and added to eppendorf containing 1.0ml LB
Left in 37C incubator for one hour, no sign of growth - spun cells down and no visible pellet, resuspended in 0.9ml and decided to leave overnight
Will plate up cells on chloramphenicol tomorrow and they should have grown by Monday.
Annoyingly, the Red/ET recombination plasmid is lost at 37C, but it was worth following the procedure to see if it actually works first time. If we need to repeat it, the plates can be grown at 30C so that there is no need to retransform with the recombination plasmid.
09/10/10
Cells left to grow for ~20 hours
Plated from 100microlitres from 900microlitre suspension onto 4 chloramphenicol plates marked JM109/lambda + cat/sacB and -cat/sacB (control) and DH5-alpha/lambda + cat/sacB and - cat/sacB (control)
Spun cells down at 7000rpm for 2 minutes - gave decent sized pellet. Removed 700microlitres of supernatant and resuspended cells
Plated last 100microlitres onto a further 4 chloramphenicol plates
Plates will be left in 37C incubator - cells were concentrated enough on second set of plates so that if they are cml resistant they will grow by tomorrow.
10/10/10
Significant growth of red colonies on DH5-alpha+ (concentrated) but none on DH5-alpha- (not-concentrated). This is highly confusing as there should be no RFP in the cells. We now believe this to be a contamination.
There is growth on both of the negative plates of the JM109 cells, but none on the positive plates. This could be a labelling error.
There is no growth on any of the other plates.
11/10/10
No growth on any of the plates that had not grown yesterday.
Colonies on JM109- plates do not smell of indole. If they are E.coli then this is promising. Subcultured 4 colonies from each plate onto cml plates.
Ran gels of PCRs performed on 07/10. No bands in the three lanes from subcultures, but a strong band from the PCR of the ligation.
12/10/10
The subcultures from yesterday have not grown particularly well. The cells are white, unlike normal E.coli, and the colonies are uncharacteristically runny. Ran PCR of subcultures to determine whether they contain the insert. (with insert ~2.5kb, without ~1.5kb)
Ran this PCR on a gel. Gave no bands. First attempt at BRIDGE did not work.
Alterations to be made on second attempt:
1- Resuspend cells in 20-50microlitres after cleaning to increase transformation efficiency
2- Grow at 30C instead of 37C after electroporation so as not to lose recombination plasmid
3- Use normal chloramphenicol plates rather than cml+iptg+xgal.
14/10/10
Prepared overnight cultures:
- DH5-alpha/lambda in 5ml LB with 1.5microlitres tet15
- JM109/lambda in 5ml LB with 5microlitres tet15
Grew cells at 30C, 200rpm
- tnaA PCR, JM109, MK2+4, CF 6.
15/10/10
Prepared secondary liquid cultures:
- 5ml LB + 1.5microlitres tet15 inoculated with 0.3ml overnight DH5-alpha/lambda culture
- 5ml LB + 5microlitres tet15 inoculated with 0.3ml overnight culture JM109/lambda
Grew cells at 30C, 200rpm for 2 hours
Transferred 1.5ml of both cultures to individual eppendorfs and spun down cells for 3 minutes at 8000rpm. Removed supernatant and added another 1.5ml. Spun down cells again and removed supernatant.
Suspended cells in 1ml cold sterile water and spun them down for 1 minute at 10000rpm before removing supernatant.
Repeated this twice more.
Resuspended cells in 100microlitires cold sterile water.
Added 40microlitres of the relevant cell culture to cold sterile electroporation cuvettes labelled J+, J-, D+ and D-.
Added 2microlitres DNA to the J+ cuvette.
Added 1microlitre DNA to the D+ cuvetter.
Prepared 8 eppendorfs, 4 containing 1.5ml LB.
Electroporated all 4 samples.
The voltage decay readings for the J-, D+ and D- cells were all above 4.0, but the reading for the J+ was 3.30. This probably indicates that there is too much DNA in the sample as the charge dissapated too quickly. From now on only 1microlitre of DNA will be used.
Immediately after electroporation, 500microlitres of LB was added to the cuvette from an eppendorf to resuspend the cells and transferred back to the eppendorf. The LB was mixed and 750microlitres were transferred to a clean eppendorf.
For each transformation, half the cells were grown at 30C overnight and half were grown at 37C overnight.
16/10/10
Spun down cells from each of the 8 eppendorfs at 8000rpm for 3 minutes and removed 600microlitres of supernatant.
Spread transformants onto chloramphenicol (one plate each) and grew at relevant temperatures for 48 hours.
18/10/10
Only growth on plates appears to be non-E.coli. Major contamination has only occurred on 30C plates. Whatever it is, it seems to grow better at 30C.
Since this is the second time we have seen this type of contamination the next attempt at the protocol will be combined with taking samples of cells at each step.
19/10/10
Made more tet15 plates, as will need about 40 this week.
Prepared overnight cultures of DH5-alpha/lambda and JM109/lamda, both grown at 30C.
20/10/10
Took a sample of each overnight culture and spread it onto two tetracycline plates (1), one grown at 30C the other grown at 37C. The reasoning here is that any contaminants at this stage must be tetracycline resistant.
Added 0.3ml overnight culture to fresh LB with tetracyline. Grew for 2 hours at 30C with shaking.
Sampled each culture again, this time plating to both chloramphenicol and tetracyline plates. (2)
NB: JM109 is not growing as well as DH5alpha in liquid culture. We may need to change the concentration of tet in one or both slightly.
Something of a mini catastrophe...I knocked over my DH5alpha liquid culture and lost half the contents. Have added LB and a proportionate amount of tet15 and left both in the 30C shaker for another hour. This should recover enough cells to continue, however it also means the LB may contaminate the DH5alpha...if it is the LB, then only DH5alpha will be contaminated.
In the mean time, am making 20-26 plates of cml.
Added 100microlitres of 20% L-arabinose to each culture and left for one hour at 37C with shaking. Prepared cold water, cuvettes and eppendorfs with LB.
Sampled cultures again, plating to chloramphenicol only. (3)
Washed cells with cold water.
Sampled cells again and plated to chloramphenicol. (4)
Electroporated two sets with the cat/sacB construct and and two without. Transferred to LB in eppendorfs for recovery and grew each set of four at both 30C and 37C for an hour.
Sampled cells again and plated to chloramphenicol. (5)
Innoculated 5ml liquid cultures with 5microlitres of cml and 600microlitres of the transformed sample and grew overnight at 30C and 37C with shaking.
21/10/10
No growth on any of the chloramphenicol plates from yesterdays samples. This suggests the usual contaminant has not yet reached the cultures. There is growth on each tetracycline plate, which is expected as the cells still contain the Red/ET plasmid at this point.
Unfortunately there is no growth in the positive chloramphenicol liquid cultures. There is growth in the D- at 37C culture which presumably is one of our contaminants. I will keep an eye on the D- 37C plate and possibly plate out the liquid culture to examine the colonies. This contamination must have occurred during the final stage of the protocol from yesterday and must be human error as it is an isolated incident.
22/10/10
Set up secondary liquid cultures in tet15 (3.5microlitres for JM109 as it seems to be struggling to grow) and left to grow in 30C shaker for 3 hours.
Plated out D- 37C cell culture that survived chloramphenicol to determine strain.
Checked chloramphenicol sample plates again:
Sample 2- White colony growth on both JM109 plates but none on DH5alpha
Sample 3- No growth (contaminants have disappeared, killed by arabinose?)
Sample 4- White colony growth 30C plates only, two micrococcal colonies on J 37C
Sample 5- A few very small white colonies on 30C plates D-, D+ and J-, no growth on D- at 37C
The bulk of the contamination appears to occur at the washing step, but whatever they are, they don't seem to be able to survive cml40 in a liquid culture. Confusingly, whatever has contaminated the D- culture at 37C doesn't seem to be on the plates. This probably means it is an isolated event. I don't know if the same goes for the contaminants in sample 2. These are more confusing as they disappear after the arabinose is added. It is possible that they do not survive either in arabinose or at 37C in liquid.
Next step to remove contaminants:
Previously the washing step had been performed in the lab away from the clean hood. From now on, any stage where the cells are exposed will take place under the clean hood.
A control will be set up at this step to confirm that contamination is occurring here and not previously. A second set of eppendorfs will be washed repeatedly with water and then 100microlitres of the water will be plated along with the actual samples.
Growth on the negative control will indicate that either the water or the eppendorfs is contaminated.
No growth on either will indicate that the use of the clean hood makes a difference.
Next step in improving the efficacy of BRIDGE:
The cat gene we're using is not very strongly induced. Couple this with the fact that DH5alpha and JM109 both struggle to grow at regular concentrations of antibiotics and we can assume that any recombinants we do get won't be able to grow in cml40. This time I will grow them overnight in a lower concentration of chloramphenicol. 3microlitres for JM109, 2microlitres for DH5alpha.
This might be the last time I will be able to attempt BRIDGE before the wiki freeze. Fingers crossed.
Maximised biomass this time by growing JM109 in 3.5microlitres tet15 and DH5alpha in 1.75microlitres as usual, for 2 and a half hours after adding 0.5ml overnight culture rather than 0.3ml.
After the induction step spun down whole cell culture and kept cells under clean hood whenever they were exposed.
Also took negative controls at washing step of sterile water and LB, both kept in eppendorfs for 10minutes.
If there is growth on:
- Sterile water only - sterile water is contaminated
- LB only - LB is contaminated
- Both - eppendorfs are contaminated
Also took samples of actual cell cultures at this step.
Electroporated cells as normal. Grew at recovery step for 2 hours.
Grew all 8 samples in cml40: JM109 in 3.5microlitres, DH5alpha in 1.75microlitres.
Grew positive transformants in tet15 (normal concentrations) to determine if the cells were still alive.
If the positive cells grow in:
- cml and tet - recombinants
- cml but not tet - contaminant
- tet but not cml - non-recombinant
- neither - cells have died
Transformed JM109 cells with pSB1A2+cat to characterise cat and Edinbrick as a negative pSB1A2 control. Plated cells to ampicillin.
23/10/10
The negative controls from the washing step have not grown, but then neither have the samples, so no contamination on anything.
There is growth in 3 of the 4 tetracyline cultures from yesterday (JM109 has not grown at 37C, possibly because of inability of plasmid to replicate). This means the cells are not, in 3 cases, being killed by the electrporation.
There is no growth in any of the negative controls, so no indication of contamination.
No growth in any of the cml positive cultures which indicates that the recombinase genes are not being induced properly.
The D- 37C contamination from Wednesday turned out to have RFP in it. This is quite confusing but, as stated earlier, all contaminations have been removed from the procedure.
Good growth of cat and Edinbrick transformants on ampicillin. Have subcultured 4 of each, again to ampicillin.
24/10/10
Subcultured the cat and Edinbrick transformants from ampicillin to cml40.
25/10/10
No significant growth on subcultures.
Set up titre of cml resistance using positive 30C transformants and negative controls. All grown in tetracycline and in increasing concentrations of chloramphenicol. Grown overnight at 30C and 200rpm.
26/10/10
Growth on Edinbrick on cml40 but not on cat. Could be mislabelling. Have re-attempted subculture from transformant plates but if initial plates are labelled wrong then the results will be the same.
Results from titre. Conclusion: recombination isn't working as cat has been known to show resistance to cml10-40.
| Ju109 (tet15 / 5ul) | DH5a (tet3.75 / 1.75ul) | |||
|---|---|---|---|---|
| Cml | + cat/sacB | control | + cat/sacB | control |
| 0 / 0 ul | + | + | + | + |
| 4 / 0.5 ul | - | - | - | - |
| 8 / 1 ul | - | - | - | - |
| 12 / 1.5 ul | - | - | - | - |
| 16 / 2 ul | - | - | - | - |
| 20 / 2.5 ul | - | - | - | - |
| 24 / 3 ul | - | - | - | - |
27/10/10
Successful growth of cat on chloramphenicol with inhibited growth of Edinbrick. Have added photo to wiki and registry for characterisation.
Chris has made an UP-kan-sacB-DOWN for the next attempt with BRIDGE. Will be using K12 as JM109 and DH5alpha are apparently not good hosts for recombination.
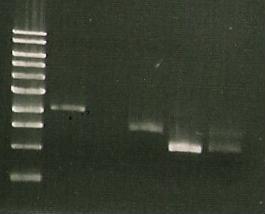
Chris also digested the Red/ET plasmid with EcoRI. This should produce 2 distinct bands (from 3 EcoRI sites). The gel below shows these bands which are the right size so we believe that the plasmid is correct.
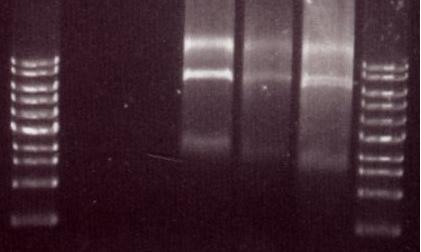
 "
"