Team:Heidelberg/Project/Capsid Shuffling/ViroBytes
From 2010.igem.org
(→ViroByte design) |
|||
| Line 70: | Line 70: | ||
==ViroByte design== | ==ViroByte design== | ||
| + | <br /> | ||
| - | + | ===Anchor=== | |
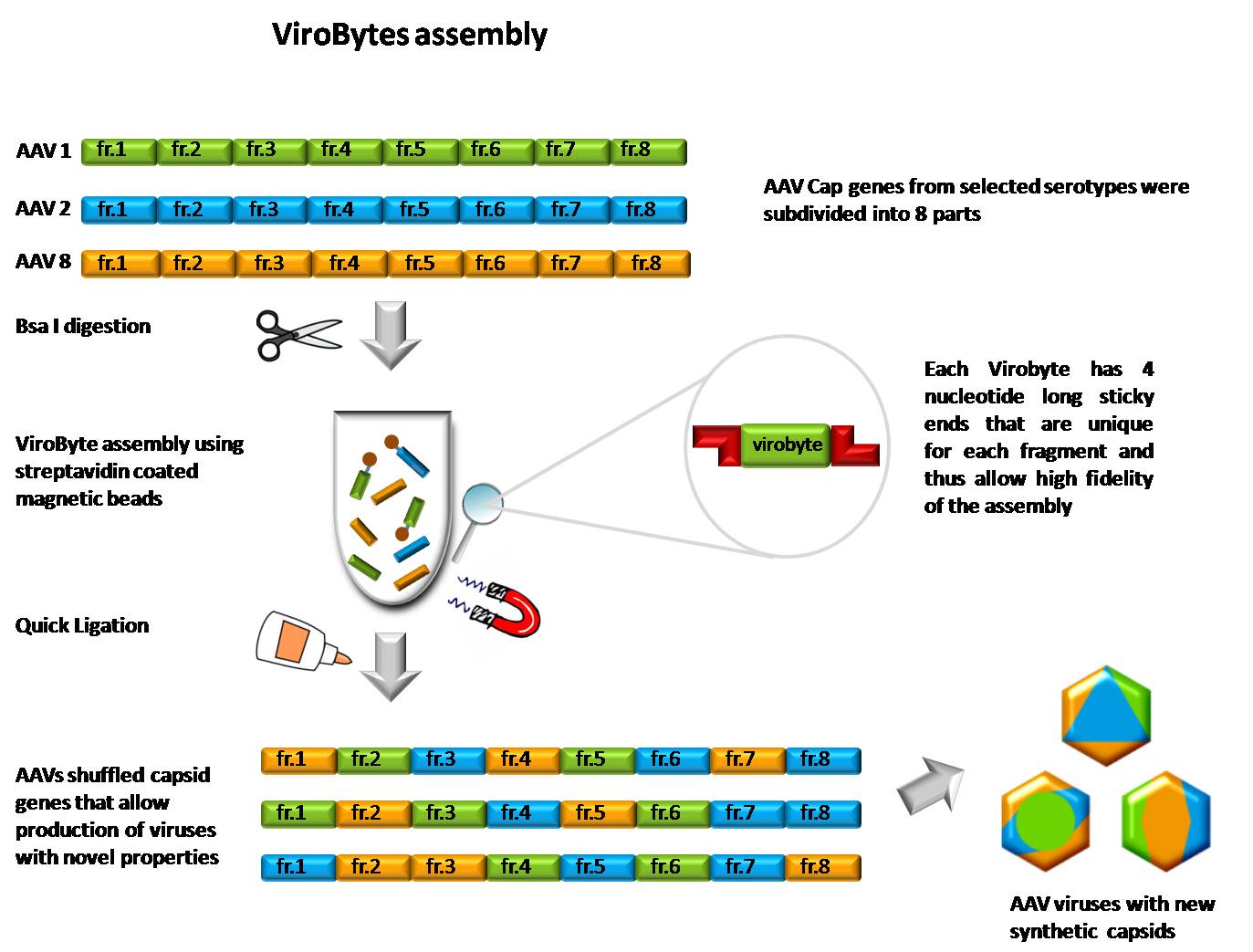
Anchor for ViroByte assembly was designed according to [https://2009.igem.org/Team:Alberta/DNAanchor BioByte Anchor System]. We used 5'-biotinylated oligos which were annealed with Anchor_complementary sequences (see [https://2010.igem.org/Team:Heidelberg/Project/Capsid_Shuffling/ViroBytes#Protocols Protocols]). The final Anchor construct contains important restriction sites as well as the VirByte_for sequence used for final PCR amplification. First recognition site is HindIII which we used for separating the beads from the assembled DNA before PCR. Second restriction sequence corresponds to PacI which is a common non-cutter within all AAV serotypes used and can be emplyed for further ligation into an appropriate vector. Most importantly the 3'-end forms a sticky end compatible with the so-called "start overhang" of all fragments'''1'''. | Anchor for ViroByte assembly was designed according to [https://2009.igem.org/Team:Alberta/DNAanchor BioByte Anchor System]. We used 5'-biotinylated oligos which were annealed with Anchor_complementary sequences (see [https://2010.igem.org/Team:Heidelberg/Project/Capsid_Shuffling/ViroBytes#Protocols Protocols]). The final Anchor construct contains important restriction sites as well as the VirByte_for sequence used for final PCR amplification. First recognition site is HindIII which we used for separating the beads from the assembled DNA before PCR. Second restriction sequence corresponds to PacI which is a common non-cutter within all AAV serotypes used and can be emplyed for further ligation into an appropriate vector. Most importantly the 3'-end forms a sticky end compatible with the so-called "start overhang" of all fragments'''1'''. | ||
Annotated sequence of anchor oligos: | Annotated sequence of anchor oligos: | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | Anchor: | ||
5'-Biotin-tctctctctctctct''aagctt'' '''gtctgagtgactagcattcg''' '''''ttaattaa'''''c+4nt(cap specific) | 5'-Biotin-tctctctctctctct''aagctt'' '''gtctgagtgactagcattcg''' '''''ttaattaa'''''c+4nt(cap specific) | ||
| - | + | *first 15nt form ssDNA linker (improves efficiency of Anchor binding onto Steptavidin-coated magnetic beads) | |
| - | + | * ''italicized'' is HindIII recognition site | |
| - | + | * '''bold''' is amplification (VirByte_for) sequence | |
| - | + | * '''''bold italicized''''' is PacI recognition site | |
| - | + | Anchor complement: | |
5'-g'''''ttaattaa''''' '''cgaatgctagtcactcagac''' ''aagctt'' | 5'-g'''''ttaattaa''''' '''cgaatgctagtcactcagac''' ''aagctt'' | ||
| - | + | * reverse complimentary strand to Anchors | |
| + | |||
| + | |||
| + | Final Anchor construct used: | ||
| + | |||
| + | 5'-Biotin-tctctctctctctctaagcttgtctgagtgactagcattcgttaattaac+4nt-3' | ||
| + | 3'-ttcgaacagactcactgatcgtaagcaattaattg-5' | ||
| + | ===Primers=== | ||
*Fragment X forward [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#ViroBytes_primers primers]: | *Fragment X forward [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#ViroBytes_primers primers]: | ||
| Line 98: | Line 110: | ||
5'-gctac'''ggtctc'''aNNNN+~20-25nt Cap specific | 5'-gctac'''ggtctc'''aNNNN+~20-25nt Cap specific | ||
| - | + | * '''bold''' is BsaI recognition sequence | |
| - | + | * NNNN = cap specific homology sequence from all cap genes (also cutting sequence of BsaI) forming the 5'-overhang | |
Revision as of 01:59, 28 October 2010

|
|
|||
 "
"