Introduction / Objective
We constructed a mathematical model to explore and characterize the operation as well as the modulation of our experimental system.
In particular, we wanted to investigate the effect of varying parameters that we could experimentally modulate on the system. Those parameters include:
- Spatial-Gradient of Inducer Concentration
- Repressor Concentration
- Ribosome Binding Site (Rate of Translation) for Product-producing Protein
In terms of our iGEM project, this model was employed to explore the effect of IPTG concentration and diffusion, lacI concentration (determined by the combination part of constitutive promoter, ribosome binding site, lacI gene, double terminator, lac promoter/operon), and the ribosome binding site (of the final product-producing enzyme) on the concentrations of all species involved - described in the following sections - and especially on Chitin Synthase and Chitin concentration and the corresponding rates.
Model Development
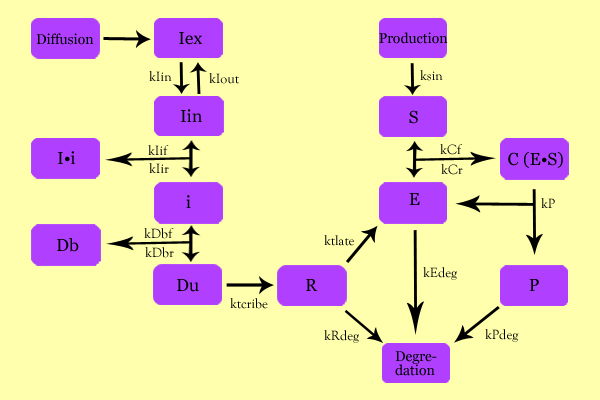
The following schematic summarizes the mathematical model we formulated to describe our system.
Variables
- Iex: External Inducer, determined by diffusion through Fick's law (IPTG in our experiment)
- Iin: Internal Inducer (IPTG)
- Ii: Inducer bound to Repressor (IPTG bound to lacI)
- i: Repressor (lacI)
- Db: Repressor-bound DNA (lacI-bound DNA(CHS3) region in plasmid)
- Dunb: transcribe-able or Repressor-unbound DNA (lacI-unbound DNA(CHS3))
- Re: mRNA for Enzyme (CHS3 mRNA)
- E: Enzyme (CHS3)
- S: Substrate (N-Acetyl Glucosamine)
- C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex)
- P: Protein Product (Chitin or (NAG)n+1)
Constants
Rate Constants:
- kIin - Inducer diffusion into cell from surrounding environment
- kIout - Inducer diffusion out of cell to surrounding environment
- kIif - Inducer and Repressor binding to form Inducer-Repressor Complex
- kIir - Inducer-Repressor Complex unbinding to form Inducer and Repressor
- kDbf - Repressor binding to DNA to form DNA-Repressor Complex
- kDbr - DNA-Repressor Complex unbinding to form free unbound DNA
- ktcribe - transcription
- ktlate - translation
- ksin - endogenous substrate production
- kCf - Substrate-Enzyme Complex Formation
- kCr - Substrate-Enzyme Complex unbinding to form Substrate and Enzyme
- kP - Substrate-Enzyme Complex forming Product and Enzyme
- kEdeg - Enzyme Degredation
- kRdeg - RNA Degredation
- kPdeg - Protein Degredation
Equations
The differential of the variables were found as follows:
- dIin/dt = kIin*Iex - kIex*Iin + kIir*Ii - kIif*Iin*i
- dIi/dt = kIif*Iin*i - kIir*Ii
- di/dt = kIir*Ii - kIif*Iin*i + kDbr*Db - kDbf*i*Dunb
- dDb/dt = kDbf*i*Dunb - kDbr*Db
- dDunb/dt = kDbr*Db - kDbf*i*Dunb
- dRe/dt = ktscribe*Dunb - kRdeg*Re
- dE/dt = ktslate*Re - kEdeg*E + kCr*CC + kP*CC - kCf*E*Si
- dSi/dt = ksin*So - kCf*E*Si + kCr*CC
- dCC/dt = kCf*E*Si - kCr*CC - kP*CC
- dP/dt = kP*CC - kPdeg*P
Assumptions
In order to determine the initial or steady state concentrations of the involved species and to determine the rate constants, the following assumptions were made:
IPTG Diffusion
First, Fick's Law of Diffusion was modeled through MATLAB. The diffusion constant used was 220um^2/s.[4]
It was assumed that IPTG was not consumed nor degraded
We also assumed that IPTG uptake was minor was compared to the concentration in the biofilm, and so that the external IPTG was determined solely by Fick's Law, not by internalization.
Constant Determination
Initially, the following initial concentration values were assumed:
- Iex: Described in the previous section
- Iin: Internal IPTG initially assumed to be 0
- Ii: 0, because no IPTG means IPTG-repressor complex
- i: Value acquired from [5] as between 10^-5 and 10^-4 and adjusted to 9.973*10^-5
- Db: Calculated, knowing the expression vector is a 200 copy number plasmid; of the DNA, approximated that 90.08% is bound
- Dunb: Calculated, knowing the expression vector is a 200 copy number plasmid; of the DNA, approximated that 9.92% is bound
- Re: Determined to be 1.82735*10^-4 by achieving steady state with the model
- E: Determined to be 6.08*10^-4 by achieving steady state with the model
- So: External Substrate calculated by using amount added in enriched media as 33.9
- Si: Determined from [3] to be 10^-1
- C: Determined to be 3*10^-5by achieving steady state with the model
- P: Determined to be 1*10^-4 by achieving steady state with the model
The rate constant values were assumed to be the following:
- kIin: .05 yielded reasonable diffusion times
- kIex: .05 since simple diffusion, the internal rate constant must be similar if not identical to the external rate constant
- kIif: .01 yielded reasonable repressor/Inducer interactions
- kIir: .01 yielded reasonable repressor/Inducer interactions
- kDbf: 100 yielded reasonable DNA/repressor interactions
- kDbr: .0011 yielded reasonable DNA/repressor interactions
- ktscribe: .01 yielded reasonable production levels
- ktslate: .01 yielded reasonable production levels
- kRdeg: .003 yielded reasonable degredation
- kEdeg: .003 yielded reasonable degredation
- kPdeg: .003 yielded reasonable degredation
- ksin: 0.000000009 yielded reasonable internalization of substrate
- kCf: .01 yielded reasonable production levels
- kCr: .01 yielded reasonable production levels
- kP: .01 yielded reasonable production levels
Results
The plots of the non-induced and the induced system are as follows:
| | Not Induced | Induced
|
| External IPTG | 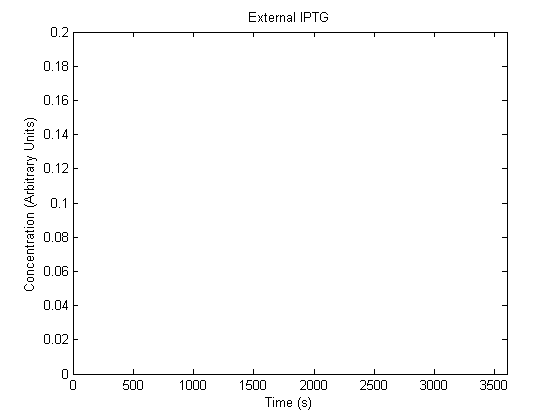 | 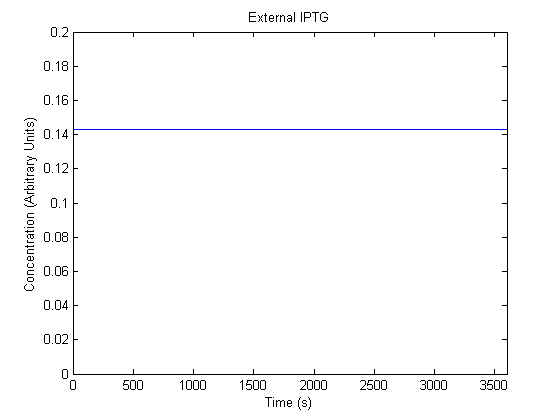
|
| Internal IPTG | 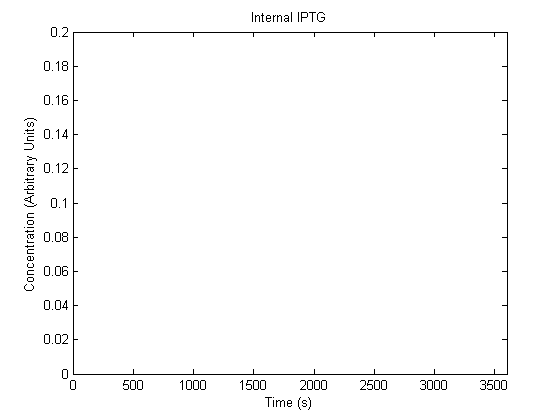 | 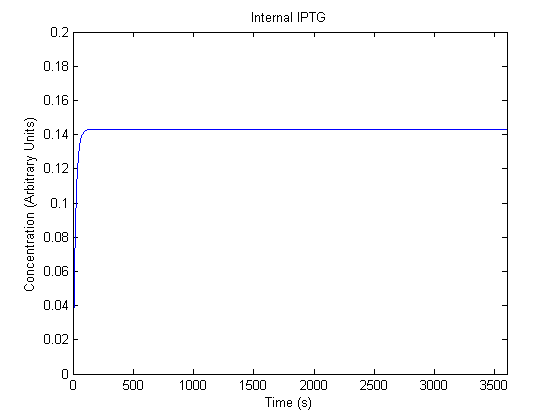
|
| IPTG bound Lac Repressor | 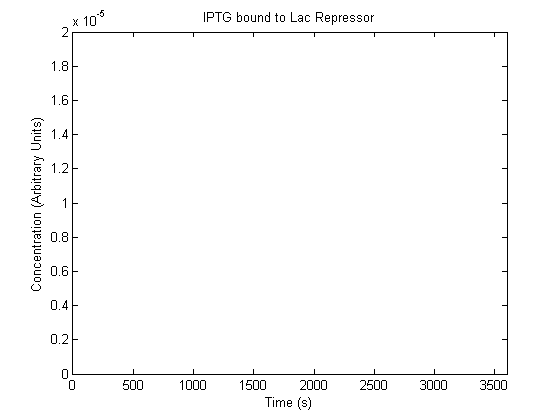 | 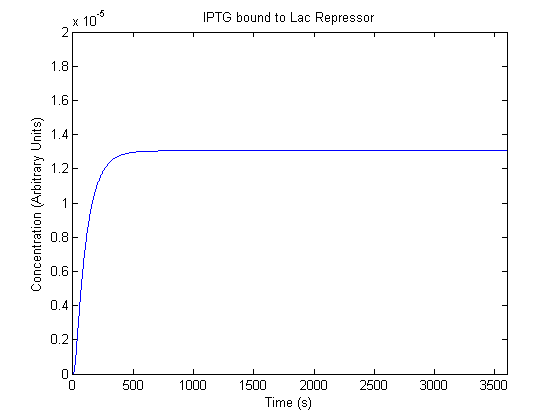
|
| Free Lac Repressor | 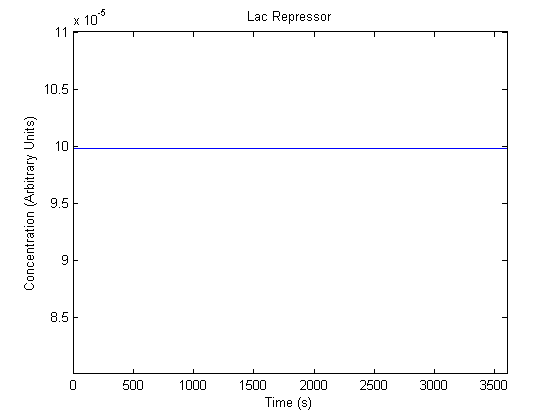 | 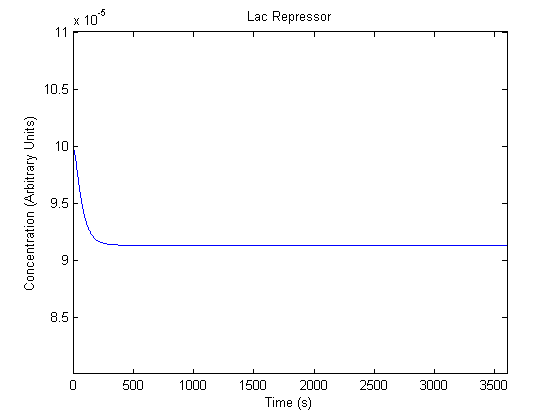
|
| Repressor Bound DNA | 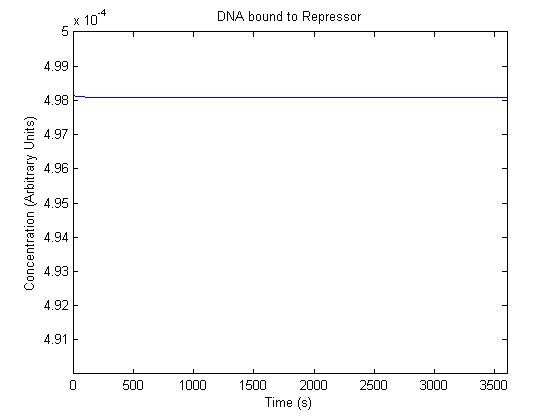 | 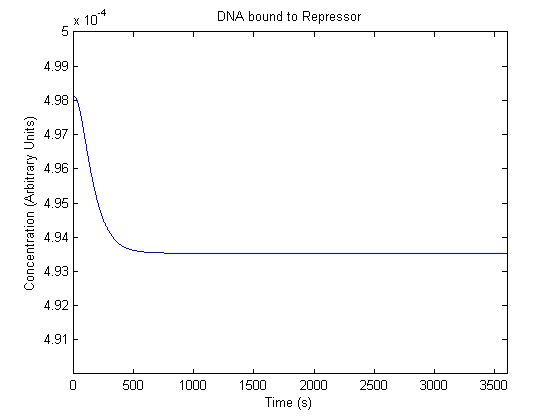
|
| Free DNA | 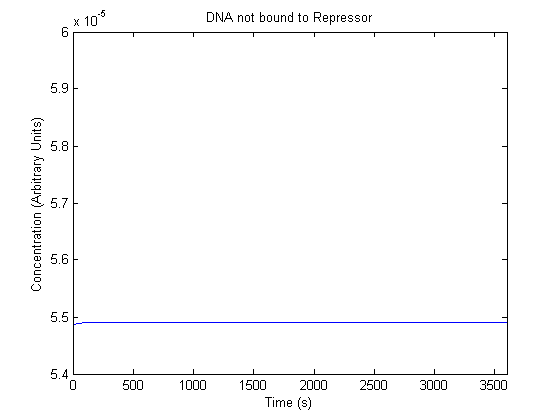 | 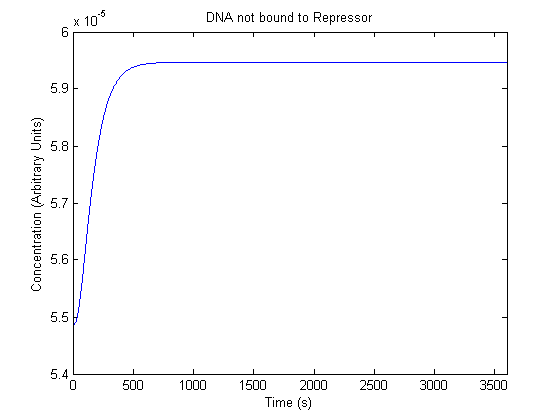
|
| mRNA | 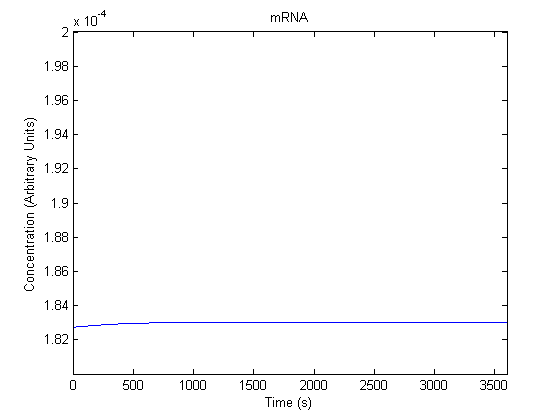 | 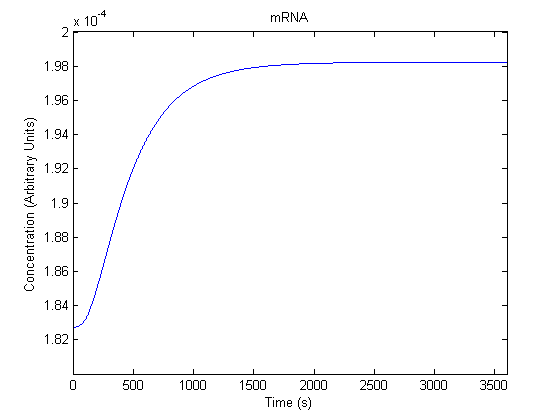
|
| Chitin Synthase 3 | 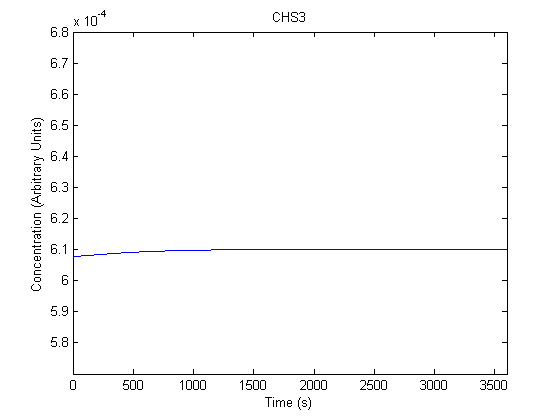 | 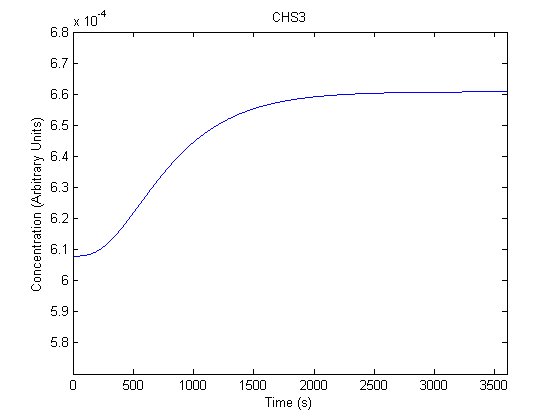
|
| N-Acetyl-Glucosamine | 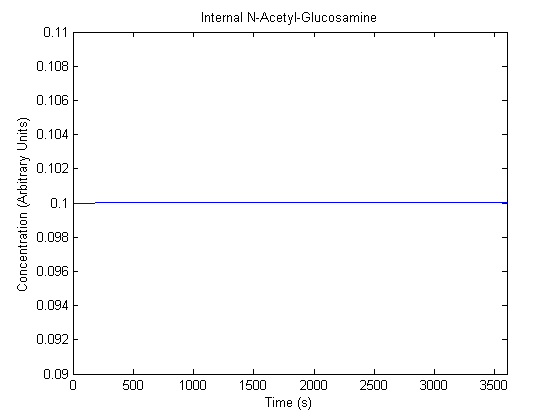 | 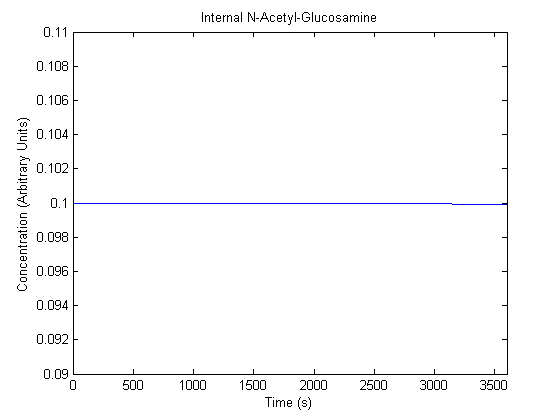
|
| CHS3-NAG/Chitin Complex | 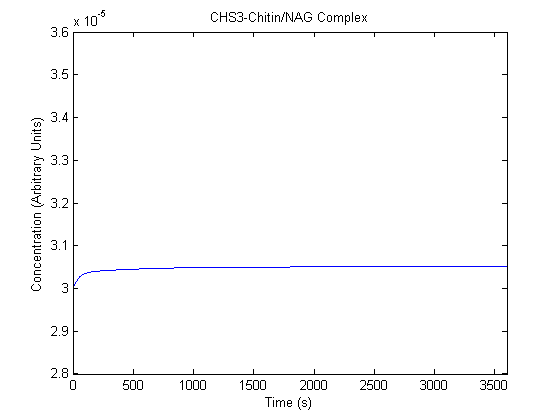 | 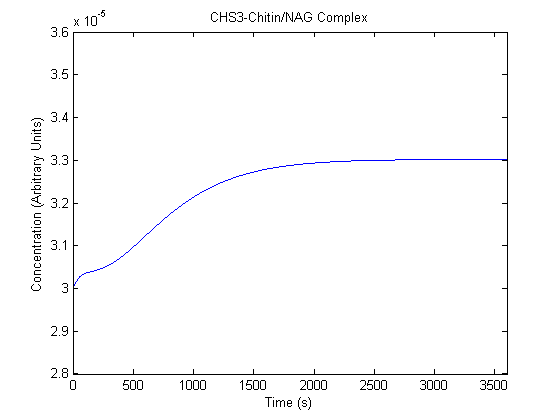
|
| Chitin | 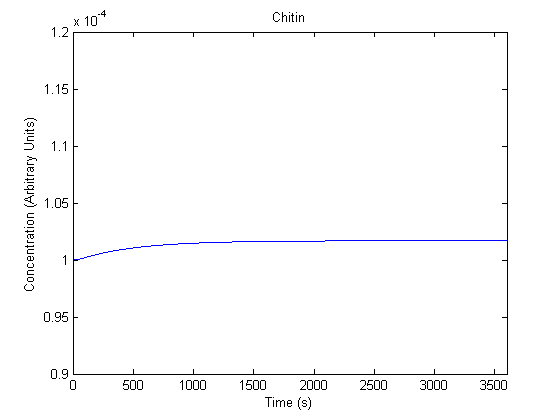 | 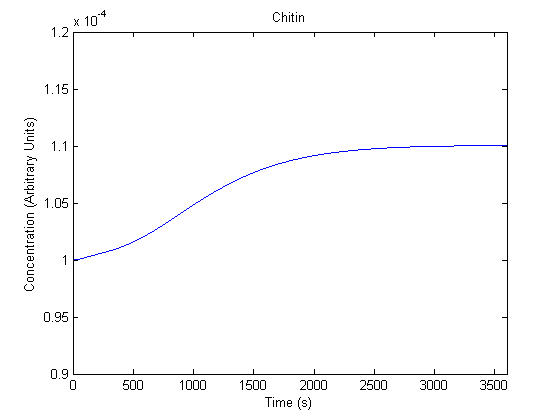
|
Future Concerns
Overall, the model, though logical, is not yet fit for application, partially due to the lack of empirical data to which the model could be fit and/or tested. Once fit, the rate constant values and initial concentration values will be much more accurate. The following concerns deal primarily with problems that could possibly not be corrected even with empirical data.
- Currently the model predicts the final product expression level between the top layer and bottom layer of a gradient-induced system as only about 0.00001%. Determine if this is primarily due to model inaccuracy or the inefficacy of diffusion-based layer differential induction method.
- Currently the model only predicts only a 10% increase in product when induced by what is suspected to be a significant concentration of inducer. This could be due to a model design problem.
- The model assumes constant substrate; this assumption may or may not be accurate.
- The model assumes all components are in first order; this assumption may or may not be sufficient.
Future Work
Aside from being generally accurate, the model should perform the following functions:
- Differentiate protein production between the top and bottom layer of the biofilm based on a diffusion-gradient inducer.
- Differentiate protein production between expression vectors with different repressor production levels.
- Differentiate protein production between expression vectors with different IPTG Induction levels.
- Differentiate protein production between expression vectors with different Promoters (k-transcribe).
- Differentiate protein production between expression vectors with different Ribosome Binding Sites (k-translate).
- Differentiate protein production between expression vectors with different Copy Number Plasmids.
References
1. A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures
William E. Bentley, Dhinakar S. Kompala
Article first published online: 18 FEB 2004
DOI: 10.1002/bit.260330108
http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf
2. Mathematical modeling of induced foreign protein production by recombinant bacteria
Jongdae Lee, W. Fred Ramirez
Article first published online: 19 FEB 2004
DOI: 10.1002/bit.260390608
http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf
3. Pool Levels of UDP N-Acetylglucosamine and UDP NAcetylglucosamine-Enolpyruvate in Escherichia coli and Correlation with Peptidoglycan Synthesis
DOMINIQUE MENGIN-LECREULX, BERNARD FLOURET, AND JEAN VAN HEIJENOORT*
E.R. 245 du C.N.R.S., Institut de Biochimie, Universit' Paris-Sud, Orsay, 91405, France
Received 9 February 1983/Accepted 15 March 1983
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf
4. Diffusion in Biofilms
Philip S. Stewart
Center for Biofilm Engineering and Department of Chemical Engineering, Montana
State University–Bozeman, Bozeman, Montana, 59717-3980
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/pdf/0965.pdf
5. Regulation of the Synthesis of the Lactose Repressor
PATRICIA L. EDELMANN' AND GORDON EDLIN
Department of Genetics, University of California, Davis, California 95616
Received for publication 21 March 1974
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC245824/pdf/jbacter00335-0105.pdf
MATLAB m-file
%IPTG PREDETERMINATION
%finite difference method
%diffusion equation (Fick's 2nd Law)
%c=c0*(erfc*x/sqrt(2*D*t))
%D*(Ci+1-2C+Ci-1)/dx^2 = Ci/dt
%x goes from 0 (top) to 100 micrometers
%D=IPTG is a modified monosaccharide, so we can estimate from Table 1 that
%its diffusion coefficient in water at 25°C will be ca. 6.5 × 10?6 cm2 s?1.
%Scaling to 37°C and taking De/Daq to be 0.25, De is found to be 2.2 × 10?6 cm2 s?1
%http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/
%%
clear
clc
%INITIALIZE
D=220; %um^2/s
c0=14.298; %mg so 3ml spray of 20mM iptg
%in comparison 23.83mg of iptg is in 5ml 20mM
dx=1; %um
xmax=100; %um; 100um total
dt=.002; %s
tmax=10; %s; 1 hour total
C=zeros(xmax/dx,tmax/dt); %rows = same time %also, (x,t) x is row, t is column
C_Rate=zeros(xmax/dt,1);
C(1,1)=c0;
%Time Step
for t=1:1:tmax/dt-1; %s
for x=1:1:(xmax/dx) %um
if x==1%account for x=1 boundary condition
C_Rate(x)=D*(C(x+1,t)-C(x,t))/(dx^2);
elseif x==xmax/dx %account for x=xmax boundary condition
C_Rate(x)=D*(C(x-1,t)-C(x,t))/(dx^2);
else%BULK
C_Rate(x)=D*(C(x-1,t)+C(x+1,t)-2*C(x,t))/(dx^2);
end
C(x,t+1)=C(x,t)+C_Rate(x)*dt;
end
t*.002
end
%%
%Compile into 1s parts
%newC=zeros(100,tmax/dt/500);
counts=1;
for t=1:500:tmax/dt-1 %.002*x=1;
newC(:,counts)=C(:,t);
counts=counts+1;
end
%%
%visualize newC
for t=1:1:tmax/dt/500
XX=0:dx:xmax-dx;
YY=newC(:,t);
plot(XX,YY,'k.');
axis([0 100 0 2])
text(50,1.8,sprintf('Time is: %g s', t*dt));
text(50,1.7,sprintf('IPTG Mass balance is: %g mg', sum(C(:,t))));
xlabel('Distance from surface (um)')
ylabel('IPTG (mg)')
Mov(t)=getframe();
%pause(1);
end
%%
%VISUALIZE
blah=1;
for t=1:7:tmax/dt
XX=xmax-dx-100:-dx:0-100;
YY=C(:,t);
plot(YY,XX,'k.');
axis([0 2 -100 0])
text(1.2,-30,sprintf('Time: %g s', t*dt));
%text(50,1.7,sprintf('IPTG Mass balance is: %g mg', sum(C(:,t))));
ylabel('Distance from surface (um)')
xlabel('IPTG (mg)')
VID(blah)=getframe(gcf);
blah=blah+1;
end
% Updated kinetics model
%{
Iex - external inducer or iptg
Iin - internal inducer or iptg
Ii - lac inhibitor / iptg complex
i - lac inhibitor
Db - DNA bound to inhibitor
Dunb - DNA not bound to inhibitor
R - mRNA
E - Enzyme Chitin Synthase
So - External Substrate - N-Acetyl Glucosamine
Si - Internal Substrate - N-Acetyl Glucosamine
CC - Enzyme Substrate Complex
P - Protein - Chitin
%}
dt=1;
tmax=3600;
dx=1;
xmax=100;
%General Array Structure: (Depth (um), Time (s))
%Depth = 0-100 micrometers
%Time = 0- 3600 seconds
preset=zeros(xmax,tmax);
Iin=preset;Ii=preset;i=preset;Db=preset;Dunb=preset;Re=preset;E=preset;Si=preset;CC=preset;P=preset;
%initial concentration
a=load('newC.mat'); %Iex is diff - assume cell intake << exist
Iex=zeros(100,3600);%a.newC();%
Iin(:,1)=0; %initially IPTG inside the cell is 0
Ii(:,1)=0; %no iptg, no Ii
i(:,1)=0.9973*10^-4; %10^-7 to 10^-8 M which is 10^-4 to 10^5 mM http://www.ncbi.nlm.nih.gov/pmc/articles/PMC245824/pdf/jbacter00335-0105.pdf
%DNA - partsregistry - 200 copy number - 200/(6.23*10^-23)/(6*10^-16
%volume)*1000 = 5.53*10^-4mM
dper=0.9008;
Db(:,1)=5.53*dper*10^-4; %say 90% is bound no idea
Dunb(:,1)=5.53*(1-dper)*10^-4; %say 10% unbound no idea
Re(:,1)=1.82735*10^-4; %initially, no RNA
E(:,1)=6.08*10^-4; %initially no enzyme
So=33.9; %1 pill per plate ~ 750mg/100ml /221.21g/mol /1000*1000*1000 is 33.9mM
Si(:,1)=10^-1; %http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf
CC(:,1)=3*10^-5; %initially 0
P(:,1)=1*10^-4; %initially 0
%rate constants
kIin=.05;%safe to say diffuses within minutes
kIex=.05;%permeability must be similar
kIif=.01;
kIir=.01;
kDbf=100;
kDbr=.0011;
ktscribe=.01;
ktslate=.01;
kRdeg=.003;
kEdeg=.003;
kPdeg=.003;
ksin=0.000000009;
kCf=.01;
kCr=.01;
kP=.01;
%Begin Loop
%FILL IN THE BLANKS
%General Array Structure: (Depth (um), Time (s))
for tt=1:1:tmax/dt-1
for xx=1:1:xmax/dx
%Predetermine diffusion
%dIex=DIF(xx,tt) + kIin*Iin(xx,tt) - kIex*Iex(xx,tt);
dIin=kIin*Iex(xx,tt) -kIex*Iin(xx,tt) +kIir*Ii(xx,tt) -kIif*Iin(xx,tt)*i(xx,tt);
dIi=kIif*Iin(xx,tt)*i(xx,tt) -kIir*Ii(xx,tt);
di=kIir*Ii(xx,tt) -kIif*Iin(xx,tt)*i(xx,tt) +kDbr*Db(xx,tt) -kDbf*i(xx,tt)*Dunb(xx,tt);
dDb=kDbf*i(xx,tt)*Dunb(xx,tt) -kDbr*Db(xx,tt);
dDunb=kDbr*Db(xx,tt) -kDbf*i(xx,tt)*Dunb(xx,tt);
dRe=ktscribe*Dunb(xx,tt) -kRdeg*Re(xx,tt);
dE=ktslate*Re(xx,tt) -kEdeg*E(xx,tt) +kCr*CC(xx,tt) +kP*CC(xx,tt) -kCf*E(xx,tt)*Si(xx,tt);
dSi=ksin*So -kCf*E(xx,tt)*Si(xx,tt) +kCr*CC(xx,tt);
dCC=kCf*E(xx,tt)*Si(xx,tt) -kCr*CC(xx,tt) - kP*CC(xx,tt);
dP=kP*CC(xx,tt) -kPdeg*P(xx,tt);
%Iex(xx,tt+1)=Iex(xx,tt)+dIex*dt;
Iin(xx,tt+1)=Iin(xx,tt)+dIin*dt;
Ii(xx,tt+1)=Ii(xx,tt)+dIi*dt;
i(xx,tt+1)=i(xx,tt)+di*dt;
Db(xx,tt+1)=Db(xx,tt)+dDb*dt;
Dunb(xx,tt+1)=Dunb(xx,tt)+dDunb*dt;
Re(xx,tt+1)=Re(xx,tt)+dRe*dt;
E(xx,tt+1)=E(xx,tt)+dE*dt;
Si(xx,tt+1)=Si(xx,tt)+dSi*dt;
CC(xx,tt+1)=CC(xx,tt)+dCC*dt;
P(xx,tt+1)=P(xx,tt)+dP*dt;
end
tt*dt/tmax*100
end
|
 "
"