Team:Heidelberg/Project/Capsid Shuffling/ViroBytes
From 2010.igem.org
(→Introduction) |
(→Introduction) |
||
| Line 10: | Line 10: | ||
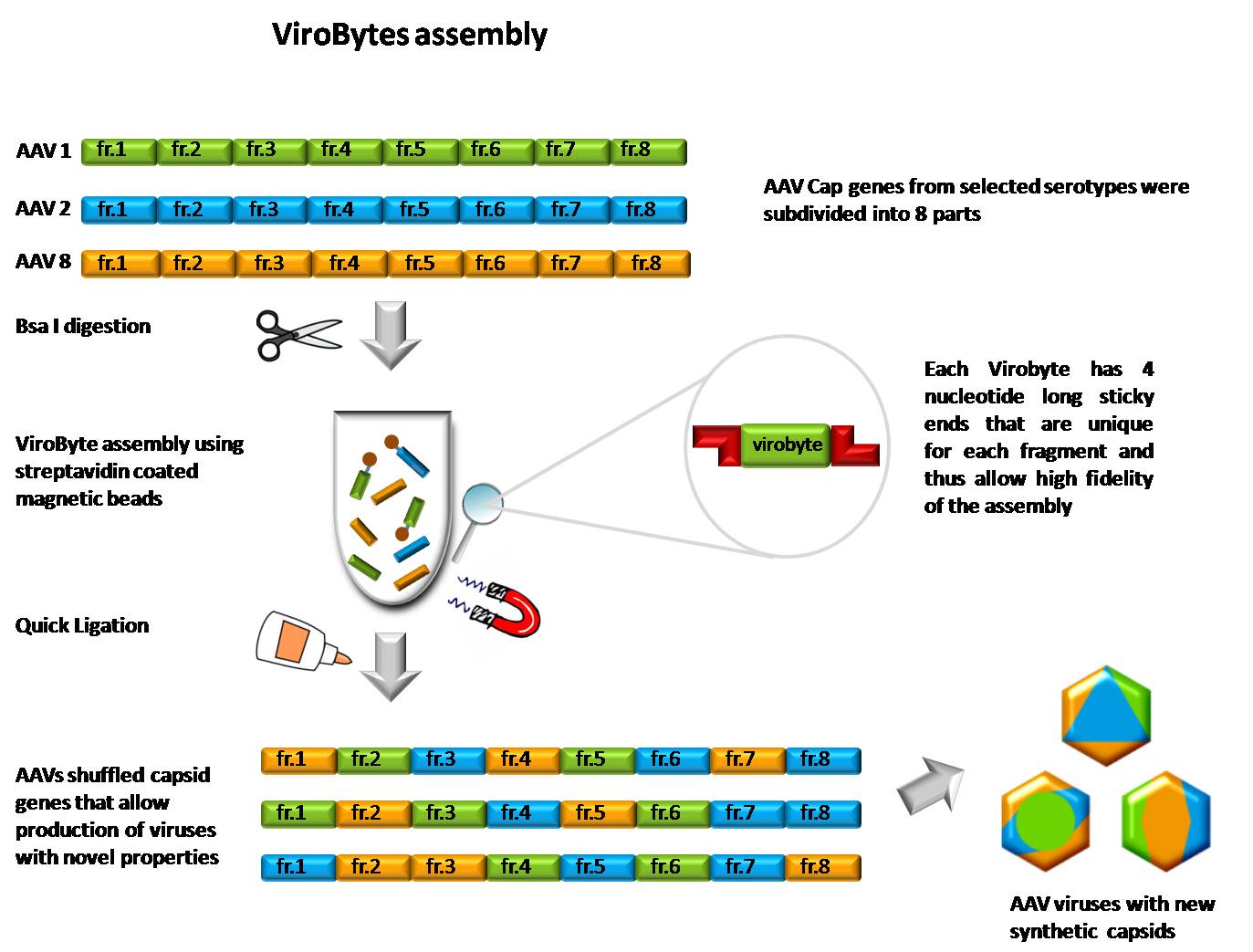
AAV serotypes 1,2,5,6,8 and 9 were selected as suitable candidates for fragmentation due to their exceptional individual properties. The analysis of Cap gene sequences revealed multiple homology regions which were then used for rational fragment formation. Total number of fragments per Cap gene is eight in our case and all fragments have similar length around ~250bp to assure similar behaviour in the ligation reaction. The Bytes are created and amplified by High-Fidelity PCR using Cap- and FragmentX-specific primers. These [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#ViroBytes_primers primers] contain flanking regions with recognition sequence for type II restriction enzyme Bsa1. Precise positioning in front of and behind the homology regions at the ends of each fragmetn ensures that the enzyme forms unique sticky ends with 4nt overhangs. This procedure allows the user to avoid arduous design of overhangs with incorporated uracils further used for USER<sup>TM</sup> digestion. Also for the purposes of shuffling, the homology regions need to be exploited in order to avoid frame shifts which cannot be easily accomplished using standard BioByte protocol. | AAV serotypes 1,2,5,6,8 and 9 were selected as suitable candidates for fragmentation due to their exceptional individual properties. The analysis of Cap gene sequences revealed multiple homology regions which were then used for rational fragment formation. Total number of fragments per Cap gene is eight in our case and all fragments have similar length around ~250bp to assure similar behaviour in the ligation reaction. The Bytes are created and amplified by High-Fidelity PCR using Cap- and FragmentX-specific primers. These [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#ViroBytes_primers primers] contain flanking regions with recognition sequence for type II restriction enzyme Bsa1. Precise positioning in front of and behind the homology regions at the ends of each fragmetn ensures that the enzyme forms unique sticky ends with 4nt overhangs. This procedure allows the user to avoid arduous design of overhangs with incorporated uracils further used for USER<sup>TM</sup> digestion. Also for the purposes of shuffling, the homology regions need to be exploited in order to avoid frame shifts which cannot be easily accomplished using standard BioByte protocol. | ||
| + | |||
| + | <br /> | ||
[[Image:Virobytes_final.jpg|thumb|350px|center|]] | [[Image:Virobytes_final.jpg|thumb|350px|center|]] | ||
Revision as of 01:02, 28 October 2010

|
|
|||
 "
"