Team:BCCS-Bristol
From 2010.igem.org
(Difference between revisions)
m (→Wetlab) |
m |
||
| Line 11: | Line 11: | ||
===Wetlab=== | ===Wetlab=== | ||
| - | + | {| | |
| + | | | ||
* '''A Well Characterised New BioBrick''' | * '''A Well Characterised New BioBrick''' | ||
| Line 26: | Line 27: | ||
:To better inform our own work, and to add knowledge to the BioBrick Registry, we have characterised Edinburgh 2009’s PyeaR BioBrick ([http://partsregistry.org/Part:BBa_K216009 BBa_ K216009]). | :To better inform our own work, and to add knowledge to the BioBrick Registry, we have characterised Edinburgh 2009’s PyeaR BioBrick ([http://partsregistry.org/Part:BBa_K216009 BBa_ K216009]). | ||
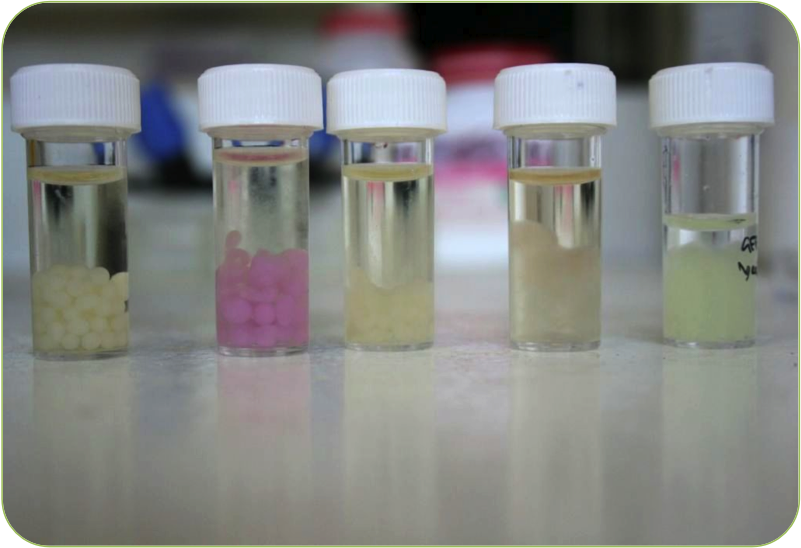
| + | |[[Image:Beads.png|thumbnail|400px|right|Our new beads]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
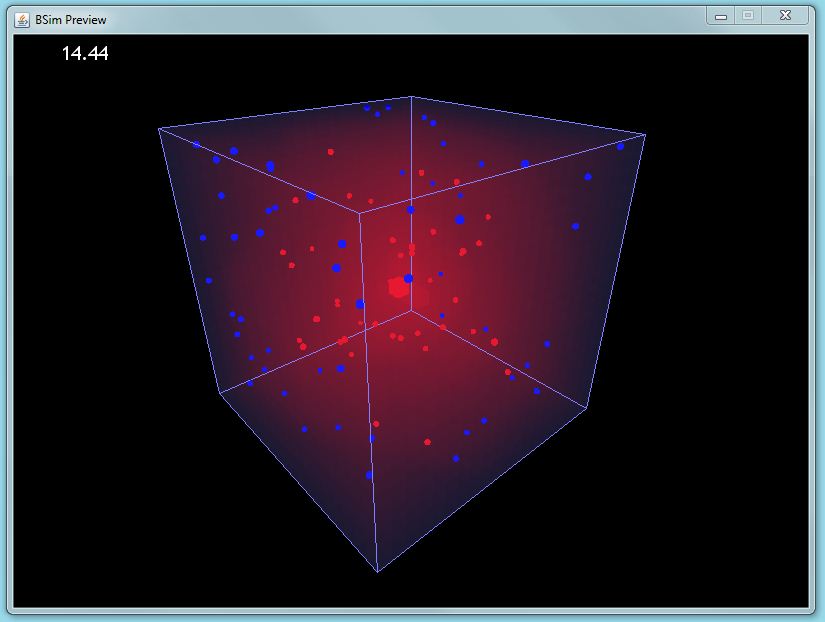
| + | |[[Image:GUI preview sample.JPG|thumbnail|400px|left|BSim screenshot]] | ||
| + | [[Image:Depth0.png|thumbnail|400px|left|Gel strand rendered in BSim]] | ||
| + | |||
| + | |||
| + | | | ||
===Modelling=== | ===Modelling=== | ||
| + | |||
*'''BSim Environmental Interactions''' | *'''BSim Environmental Interactions''' | ||
| Line 42: | Line 53: | ||
:In support of our human practices work, our modelling team have looked into the cost to farmers of using agrEcoli, and how much money and fertiliser they can expect to save. | :In support of our human practices work, our modelling team have looked into the cost to farmers of using agrEcoli, and how much money and fertiliser they can expect to save. | ||
| + | |||
| + | |} | ||
===Human Practices=== | ===Human Practices=== | ||
Revision as of 00:03, 28 October 2010
BCCS-Bristol
iGEM 2010
iGEM 2010
Achievements
Wetlab
|
|
|
Modelling
|
Human Practices
- Publicising agrEcoli
- Our new approach to human practices, building on previous work by iGEM teams, is a publicity campaign. By presenting our prototype as a functioning and marketable product, we've framed a hypothetical situation in which our project could be released.
 "
"