Team:Newcastle/25 June 2010
From 2010.igem.org
(Difference between revisions)
RachelBoyd (Talk | contribs) (→Polymerase Chain Reaction protocol) |
RachelBoyd (Talk | contribs) (→Polymerase Chain Reaction protocol) |
||
| Line 31: | Line 31: | ||
#Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase. | #Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase. | ||
| - | [[Image:fri-gel.png| | + | {| |
| + | |[[Image:fri-gel.png|300px|thumb]] | ||
| + | |[[Image:Newcastle gel 20100625.jpg|300px|thumb]] | ||
| + | |} | ||
Revision as of 14:34, 12 July 2010

| |||||||||||||
| |||||||||||||
Friday
Aims
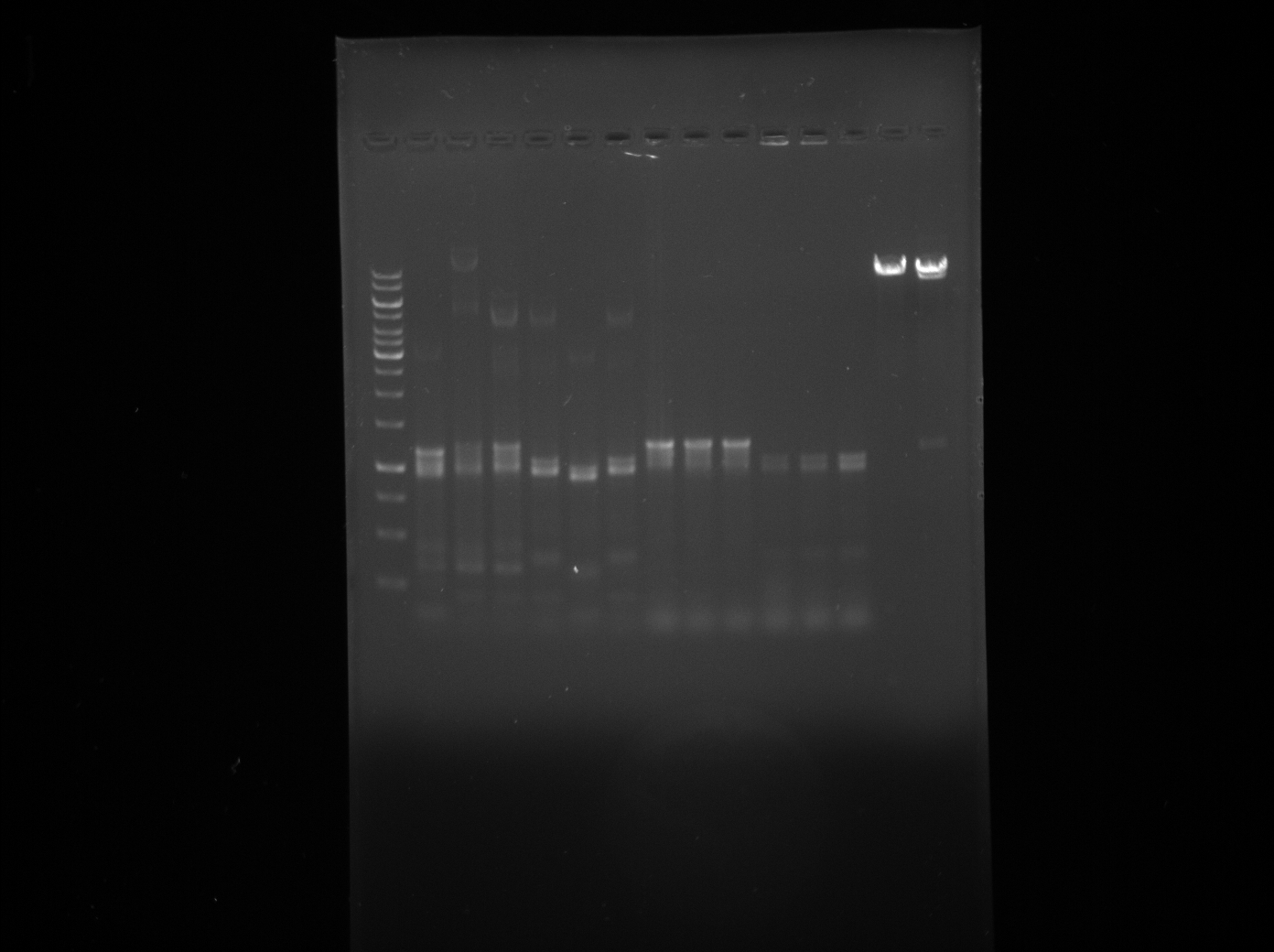
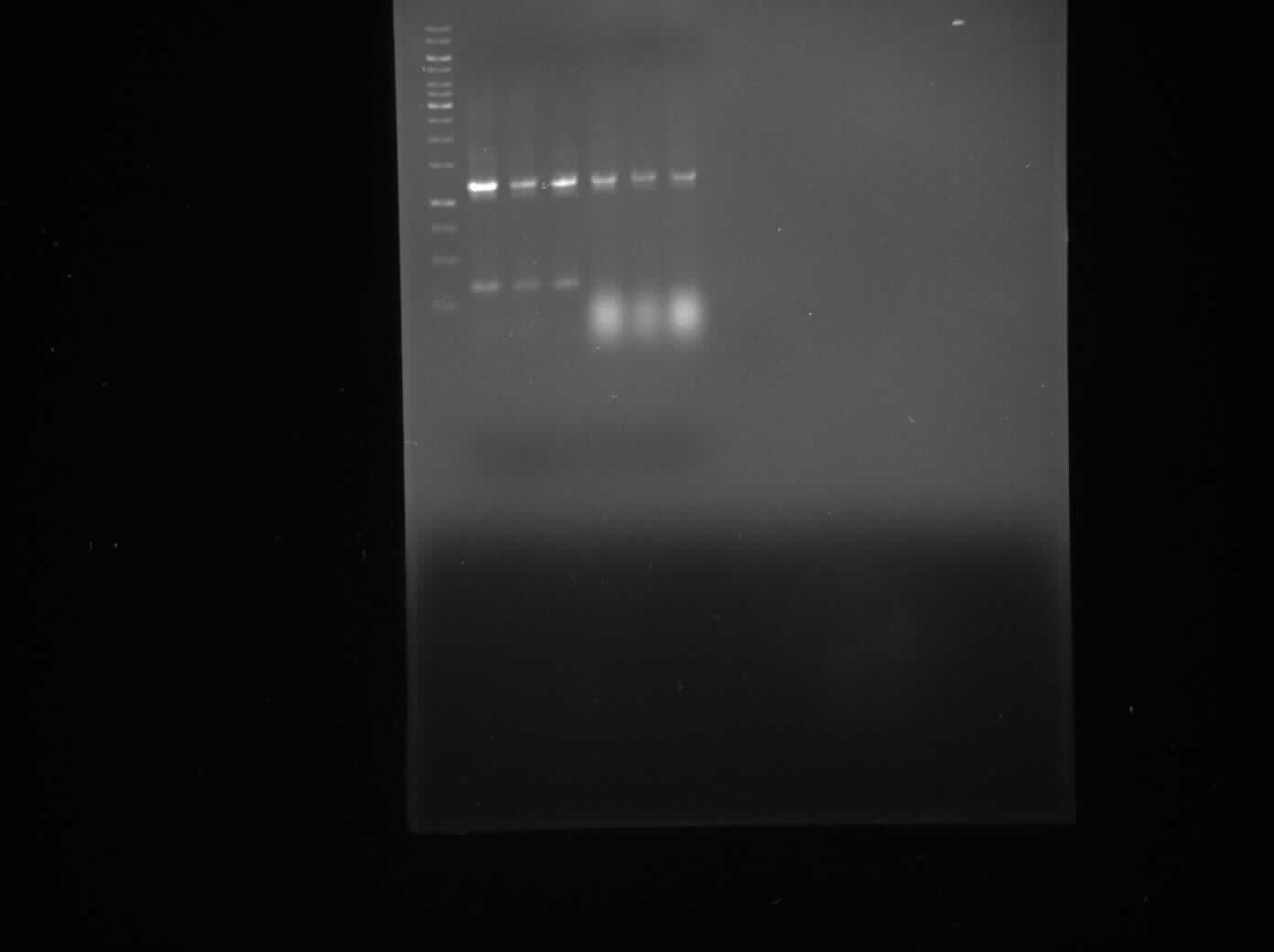
The aim of today's Lab practice is to run the products of a PCR reaction on a gel to visualise the inserts and backbones as they separate due to difference in mass.
Equipment List
- Solutions/ enzymes for PCR protocol
- GoTaq polymerase
- Gloves
- Agarose gel
- Primers(Forward and Results)
- dNTPs
- Buffers
- Ice
- Sample DNA
- Thermocycler
Polymerase Chain Reaction protocol
Polymerase chain reaction (PCR) is a technique used to amplify a single or few a copies of a piece of DNA generating thousands to millions of copies of a particular DNA sequence.
- To avoid contamination, wear gloves
- To achieve optimum results, always do everything on ice
A basic PCR set up requires several components and reagents.These components include:
- DNA template that contains the DNA region (target) to be amplified.
- Two primers that are complementary to the 3' (three prime) ends of each of the sense and anti-sense strand of the DNA target.
- Taq polymerase or another DNA polymerase with a temperature optimum at around 70 °C.
- Deoxynucleoside triphosphates, the building blocks from which the DNA polymerases synthesizes a new DNA strand.
- Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase.
 "
"