Team:Heidelberg/Project/Mouse Infection
From 2010.igem.org
(→Abstract) |
(→Introduction) |
||
| Line 21: | Line 21: | ||
Therefore AAV capsids are shuffled and under a stringent selection pressure the perfect tissue specific virus with a shuffled capsid gene should be picked. Even though AAVs are the perfect viruses in the context of gene therapy, as the vector is neither pathogenic nor toxic in cell culture and in vivo ([https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection#References Grimm, 2002]) it is still not possible to selectively choose an AAV serotype which are tissue specific. It is for example known that a lot of rational modifications of AAV capsid genes (by inserting peptides or introducing some point mutations) may lead to viruses which are really efficient in cell culture but would never work in vivo. Therefore the capsid shuffling introduces a perfect method for constructing AAV capsid genes which can be shuffled in any combination of choice. ([https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection#References Grimm et al., 2008]) <br> | Therefore AAV capsids are shuffled and under a stringent selection pressure the perfect tissue specific virus with a shuffled capsid gene should be picked. Even though AAVs are the perfect viruses in the context of gene therapy, as the vector is neither pathogenic nor toxic in cell culture and in vivo ([https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection#References Grimm, 2002]) it is still not possible to selectively choose an AAV serotype which are tissue specific. It is for example known that a lot of rational modifications of AAV capsid genes (by inserting peptides or introducing some point mutations) may lead to viruses which are really efficient in cell culture but would never work in vivo. Therefore the capsid shuffling introduces a perfect method for constructing AAV capsid genes which can be shuffled in any combination of choice. ([https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection#References Grimm et al., 2008]) <br> | ||
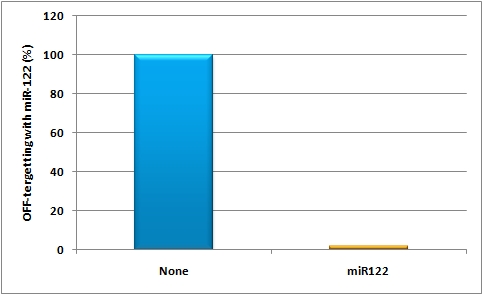
The possibility of targeting and even tuning expression in a certain tissue or cell line by using miRNA binding sites leads to even fine-tuned and more specific expression of the GOI. The binding sites can be either constructed by the miBS designer or by rational design. They can be validated using the miMeasure plasmid or the miRtuning construct. After having characterized the binding sites of choice a specific and tuned expression cassette can be introduced into a viral construct and infected into mice. And this is exactly what we did:<br> | The possibility of targeting and even tuning expression in a certain tissue or cell line by using miRNA binding sites leads to even fine-tuned and more specific expression of the GOI. The binding sites can be either constructed by the miBS designer or by rational design. They can be validated using the miMeasure plasmid or the miRtuning construct. After having characterized the binding sites of choice a specific and tuned expression cassette can be introduced into a viral construct and infected into mice. And this is exactly what we did:<br> | ||
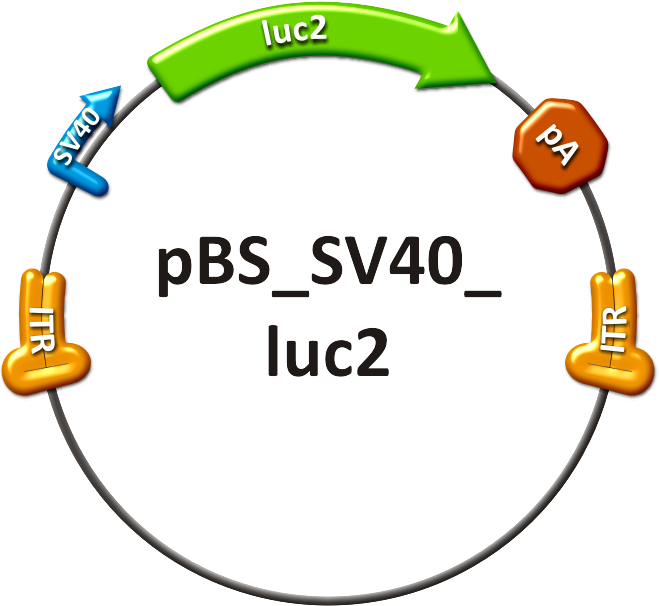
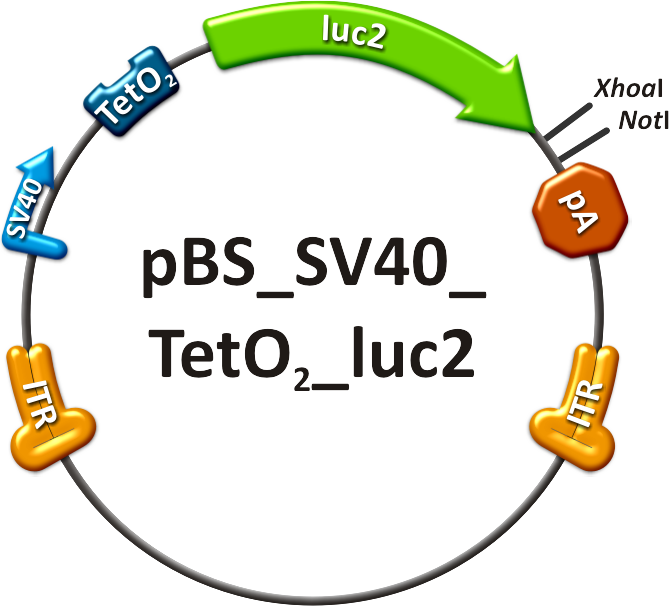
| - | We rationally designed binding sites, cloned them into the | + | We rationally designed binding sites, cloned them into the miTuner kit and characterized them. We produced and own shuffled capsid library and after applying selection pressure one clone showed already very fast good infection efficiencies in culture. For this very reason we tested our shuffled capsid and also an AAV8 WT capsid packaging a Luciferase construct driven by the same promoter. This constructs tagged with different binding site combinations of choice in the 3’UTR were used for producing viral constructs for testing on- off and tuning properties of our system. |
==Results== | ==Results== | ||
Revision as of 21:13, 27 October 2010

|
|
|||
 "
"