Team:Imperial College London/Parts/Favourites
From 2010.igem.org
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
{{:Team:Imperial_College_London/Templates/PartsHeader}} | {{:Team:Imperial_College_London/Templates/PartsHeader}} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|XylE - catechol 2,3-dioxygenase from P.putida with terminator | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|[http://partsregistry.org/Part:BBa_K316003 '''BBa_K316003'''] | ||
| + | |- | ||
| + | |Catechol or catechol 2,3-dioxygenases (C2,3O) + O(2) is converted by a ring cleavage into 2-hydroxymuconate semialdehyde which is the toxic and bright yellow-coloured substrate<cite>1</cite>. This is a key enzyme in many (soil) bacterial species used for the degradation of aromatic compounds. Catechol 2,3-dioxygenase<cite>2</cite> was originally isolated from Pseudomonas putida and is a homotetramer of C230 monomers. The tetramerization interactions position a ferrous ion critical for enzymatic activity. It has been deduced that intersubunit interaction is essential to produce a functioning enzyme after performing N and C terminal modifications on the monomer. Coming together the subunits generate an active site. The reaction itself takes place within seconds after the addition by Pasteur pipette or spraying of catechol at a 100mM stock solution diluted with DDH20 (used by our lab.) The toxic byproduct is thought to interfere with cell wall integrity and cellular machinery such that exposed cells gradually die. | ||
| + | |||
| + | |||
| + | '''Safety''' | ||
| + | |||
| + | Catechol is classed as irritant in the EU but as toxic in the USA, as well as being a possible carcinogen. It should therefore be handled with care and proper safety equipment. More information is available on the Material Safety Data Sheet[http://www.sciencelab.com/msds.php?msdsId=9927131]. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'><big>'''Sequence and Features'''</big></span> | ||
| + | |||
| + | <partinfo>BBa_K316003 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K316004 parameters</partinfo> | ||
| + | <!-- --> | ||
| + | |||
| + | <span class='h3bb'><big>'''Part Characterisation'''</big></span> | ||
| + | |||
| + | The enzymatic reaction catalysed by XylE can also serve as a very useful reporter. The substrate - catechol is colourless. However within seconds of its addition, the colonies or liquid cultures of XylE-expressing cells become clearly yellow<cite>3</cite> to the naked eye. This reaction allows direct measurement of XylE activity by measuring product concentrations, which absorbs light in the visible spectrum. The spectrophotometric assay compared the spectra of two cultures of E.coli (one XylE gene transformed and the other not) were compared on addition of 0.1mM Catechol substrate. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:SpectraXylE.PNG|center|500px]] | ||
| + | |||
| + | |||
| + | '''Figure I.''' XylE assay : Peak absorbance of catechol breakdown product (2-hydroxymuconic semialdehyde). | ||
| + | |||
| + | A spectrophotometric assay of two cultures of E.coli (Blue: contains <bbpart>BBa_K316004</bbpart>, Red: not expressing XylE ) were compared on addition of 0.1mM Catechol substrate. The spectra show that in XylE transformed cells, a broad peak appears at about 380nm. The absorbance at this particular wavelength is due to the yellow product of the reaction (2-hydroxymuconic semialdehyde (HMS)). | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | Characterisation data was obtained using GFP-XylE constructs <bbpart>BBa_K316007</bbpart> and XylE under two different promoters: ''B. subtilis'' derived Pveg <bbpart>BBa_K316005</bbpart> and J23101 <bbpart>BBa_K316004</bbpart> from ''E. coli''. These are described on our wiki[https://2010.igem.org/Team:Imperial_College_London/Results] and the aforementioned parts pages. | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/10368270 Kita ''et al.'' 1999] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/12519074 Okuta ''et al.'' 2003] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/6405380 Zukowski ''et al.'' 1983] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|B. subtilis transformation vector with LacI, targets amyE locus | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|[http://partsregistry.org/Part:BBBa K316027 '''BBBa K316027'''] | ||
| + | |- | ||
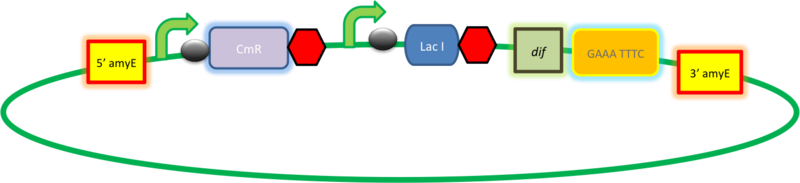
| + | |This vector has been designed using the amyE 5' and 3' integration sequences for integration into B.subtilis genome | ||
| + | |||
| + | |||
| + | '''AmyE locus''' | ||
| + | This vector has been designed using the amyE 5' <bbpart>BBa_K143008</bbpart> and 3 <bbpart>BBa_K143009</bbpart>' integration sequences for integration into ''B. subtilis'' genome. Insertion into the amyE locus provides a selection marker as the bacterium will no longer be able to breakdown starch. An iodine assay can be used to confirm integration. This phenotype makes the transformed bacterium considerably less likely to survive in natural environments. | ||
| + | |||
| + | |||
| + | '''LacI''' | ||
| + | With constitutivly expressed LacI <bbpart>BBa_K143033</bbpart> the vector is suitable for B. subtilis transformations with hyper-spank <bbpart>BBa_K143015</bbpart> controlled gene-circuits. | ||
| + | |||
| + | '''Chloramphenicol Resistance''' | ||
| + | This vector also contains a positive selection marker, flanked by two dif sites. Chloramphenicol acetyltransferase <bbpart>BBa_J31005</bbpart> provides resistance to chloramphenicol antibiotic. | ||
| + | |||
| + | '''Blunt end cloning site''' | ||
| + | PmeI restriction site <bbpart>BBa_K316013</bbpart> is designed as a cloning site. Due to the 8bp recognition sequence it is a rare site that can be used to cut the vector only once. | ||
| + | |||
| + | |||
| + | [[Image:VectorLacI.PNG|center|800px]] | ||
| + | |||
| + | '''Figure I.''' Complete amyE vector for ''B. subtilis'' genome integration with LacI expression. | ||
| + | |||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'><big>'''Sequence and Features'''</big></span> | ||
| + | <partinfo>BBa_K316027 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | |||
| + | For more information about our project please visit our wiki [https://2010.igem.org/Team:Imperial_College_London] or take the tour [https://2010.igem.org/Team:Imperial_College_London/Tour/Page_One] to learn more about the project. | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K316027 parameters</partinfo> | ||
| + | <!-- --> | ||
| + | |||
| + | |||
| + | |||
| + | |||
{| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:left;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
Revision as of 20:57, 27 October 2010
| Parts | Favourites | Full List |
| We have contributed 23 parts to the Registry of Standard Biological Parts and we really hope that other people will find them useful in the future. Here you can find detailed information on our favourite parts, or the registry information on the complete parts list. | |
| XylE - catechol 2,3-dioxygenase from P.putida with terminator | ||||||
| [http://partsregistry.org/Part:BBa_K316003 BBa_K316003] | ||||||
| Catechol or catechol 2,3-dioxygenases (C2,3O) + O(2) is converted by a ring cleavage into 2-hydroxymuconate semialdehyde which is the toxic and bright yellow-coloured substrate1. This is a key enzyme in many (soil) bacterial species used for the degradation of aromatic compounds. Catechol 2,3-dioxygenase2 was originally isolated from Pseudomonas putida and is a homotetramer of C230 monomers. The tetramerization interactions position a ferrous ion critical for enzymatic activity. It has been deduced that intersubunit interaction is essential to produce a functioning enzyme after performing N and C terminal modifications on the monomer. Coming together the subunits generate an active site. The reaction itself takes place within seconds after the addition by Pasteur pipette or spraying of catechol at a 100mM stock solution diluted with DDH20 (used by our lab.) The toxic byproduct is thought to interfere with cell wall integrity and cellular machinery such that exposed cells gradually die.
Catechol is classed as irritant in the EU but as toxic in the USA, as well as being a possible carcinogen. It should therefore be handled with care and proper safety equipment. More information is available on the Material Safety Data Sheet[http://www.sciencelab.com/msds.php?msdsId=9927131].
<partinfo>BBa_K316003 SequenceAndFeatures</partinfo>
Part Characterisation The enzymatic reaction catalysed by XylE can also serve as a very useful reporter. The substrate - catechol is colourless. However within seconds of its addition, the colonies or liquid cultures of XylE-expressing cells become clearly yellow3 to the naked eye. This reaction allows direct measurement of XylE activity by measuring product concentrations, which absorbs light in the visible spectrum. The spectrophotometric assay compared the spectra of two cultures of E.coli (one XylE gene transformed and the other not) were compared on addition of 0.1mM Catechol substrate.
A spectrophotometric assay of two cultures of E.coli (Blue: contains <bbpart>BBa_K316004</bbpart>, Red: not expressing XylE ) were compared on addition of 0.1mM Catechol substrate. The spectra show that in XylE transformed cells, a broad peak appears at about 380nm. The absorbance at this particular wavelength is due to the yellow product of the reaction (2-hydroxymuconic semialdehyde (HMS)).
Characterisation data was obtained using GFP-XylE constructs <bbpart>BBa_K316007</bbpart> and XylE under two different promoters: B. subtilis derived Pveg <bbpart>BBa_K316005</bbpart> and J23101 <bbpart>BBa_K316004</bbpart> from E. coli. These are described on our wiki[1] and the aforementioned parts pages.
References[http://www.ncbi.nlm.nih.gov/pubmed/10368270 Kita et al. 1999] [http://www.ncbi.nlm.nih.gov/pubmed/12519074 Okuta et al. 2003] [http://www.ncbi.nlm.nih.gov/pubmed/6405380 Zukowski et al. 1983]
|
 "
"