Team:Imperial College London/Strategy
From 2010.igem.org
| Line 3: | Line 3: | ||
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Assembly Strategy | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Assembly Strategy | ||
|- | |- | ||
| - | |'''Here is a quick guide to our assembly strategy. The diagram at the bottom has most of the plan; you might need to click through and download it to see all the details. The [[Team:Imperial_College_London/Parts | parts]] page has more specifics on the parts themselves.''' | + | |'''Here is a quick guide to our assembly strategy. The diagram at the bottom has most of the plan; you might need to click through and download it to see all the details. The [[Team:Imperial_College_London/Parts | parts]] page has more specifics on the parts themselves, or follow the [[Team:Imperial_College_London/Lab_Diaries | lab diaries]] to see our progress.''' |
|} | |} | ||
{| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
Revision as of 17:15, 27 October 2010
| Assembly Strategy |
| Here is a quick guide to our assembly strategy. The diagram at the bottom has most of the plan; you might need to click through and download it to see all the details. The parts page has more specifics on the parts themselves, or follow the lab diaries to see our progress. |
| Overview |
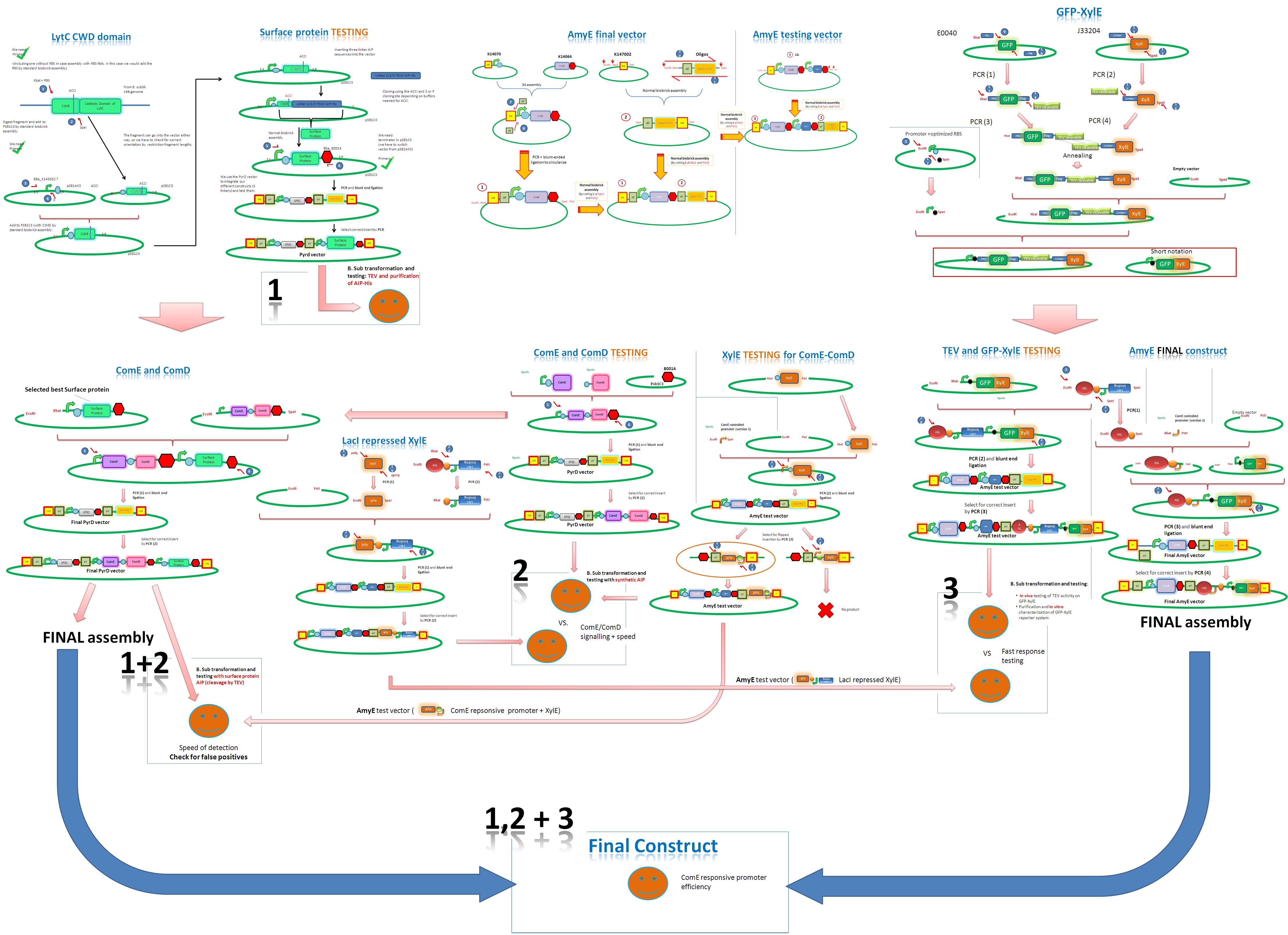
| To optimize the construction of our system, we finalized the complete assembly strategy ad hoc.
The three main streams of assembly were:
These would then be combined with the constructs of ComE, ComD and TEV (which we submitted for synthesis) in order to complete the gene circuit. 1. Assembly of the Surface Protein Construct We isolated the Cell-wall-binding domain of the LytC protein from the genome of B. subtilis. After introduction of an optimized RBS and the constitutive PVeg promoter we completed the assembly with the addition of the different versions of linker-AIP constructs which we received from synthesis. 2. Assembly of the AmyE and PyrD vectors The vectors were assembled by analogous strategies using parts K14070, K14064, K147002, 12L and several synthesized sequences. Next to the final version, a version of the AmyE vector, containing a LacI gene, was assembled, in order to carry out controlled characterization experiments with LacI repressed genes. 3. Assembly of the GFP-XylE Construct The sequence encoding the GFP-XylE fusion protein was assembled by progressive addition of sequence parts using PCR. The final step was the introduction of the optimized RBS and promoter sequences was performed by standard assembly. In order to prevent transcriptional read-through into sequences to the expression of which our system is highly sensitive, we assembled flipped versions of the LacI-TEV-protease. Introduced by the AmyE-vector, the LacI-TEV-protease sequence is therefore insulated from the influence of genes adjacent to the locus of integration. Both a LacI-inducible and a ComE-inducible construct of TEV-protease were assembled and combined with the GFP-XylE fusion protein sequence in the appropriate AmyE vector versions. A flipped version of XylE was assembled as testing construct to use for standardization during characterization of the GFP-XylE fusion protein in vivo. Finally, in order to ensure optimal molar relations of response regulator to sensory histidine kinase, we assembled ComE and ComD with respectively optimized ribosome binding sites, as under the influence of a single PveG promoter. These were, in sequence with the surface-protein gene, introduced into the integration locus via the PyrD vector. |
 "
"