Team:BCCS-Bristol
From 2010.igem.org
| Line 1: | Line 1: | ||
{{:Team:BCCS-Bristol/Header}} | {{:Team:BCCS-Bristol/Header}} | ||
__NOTOC__ | __NOTOC__ | ||
| - | |||
| - | |||
| - | |||
<center> | <center> | ||
[[Image:AgrEcoli_bead_logo.png|upright= 3.3|frameless|center]] | [[Image:AgrEcoli_bead_logo.png|upright= 3.3|frameless|center]] | ||
Revision as of 17:01, 27 October 2010
iGEM 2010
Project Abstract
Motivation
Many crop types need to be harvested and replanted from scratch on an annual basis. To maintain nutrient levels in the soil, farmers often have to put down considerable quantities of fertiliser. This is costly, and a fair proportion of it ends up landing on soil that has enough nutrients anyway. Additionally, some of the nutrients in the fertiliser are often washed away, affecting local ecosystems. This process is called eutrophication. This is a major environmental concern, as it can cause algal blooms that drain oxygen out of rivers and lakes, killing fish and other wildlife. Fertiliser production is also a significant contributor to global greenhouse emissions. Reducing excess fertiliser use would be of great benefit to both food producers and the environment.
Our Solution
Our lab has developed and characterised a cheap, versatile soil fertility sensor based on an E.coli chassis. It expresses fluorescent signals upon nutrient detection, producing a high-resolution nutrient distribution map of arable land. The ratio of two fluorescent signals allows farmers to quantify soil nutrient content. agrEcoli bacteria, encapsulated within a gel container to improve visibility and prevent escape, have been shown to work on soil in lab conditions. We have explored the marketing of our device, considering public perceptions of synthetic biology. BSim, our prize-winning modelling framework, has been extended to analyse our new biobricks’ behaviour within gel capsules. In addition, a new interface for BSim has improved its accessibility to the wider synthetic biology community, facilitating collaboration. agrEcoli optimises fertiliser use, saving farmers' money and reducing environmental damage.
How does agrEcoli work?
| Imagine viewing a cross section of the soil in a field. The topsoil is usually depleted in nutrients. Before the fertilisation process begins, the crops are harvested, leaving open soil. | |
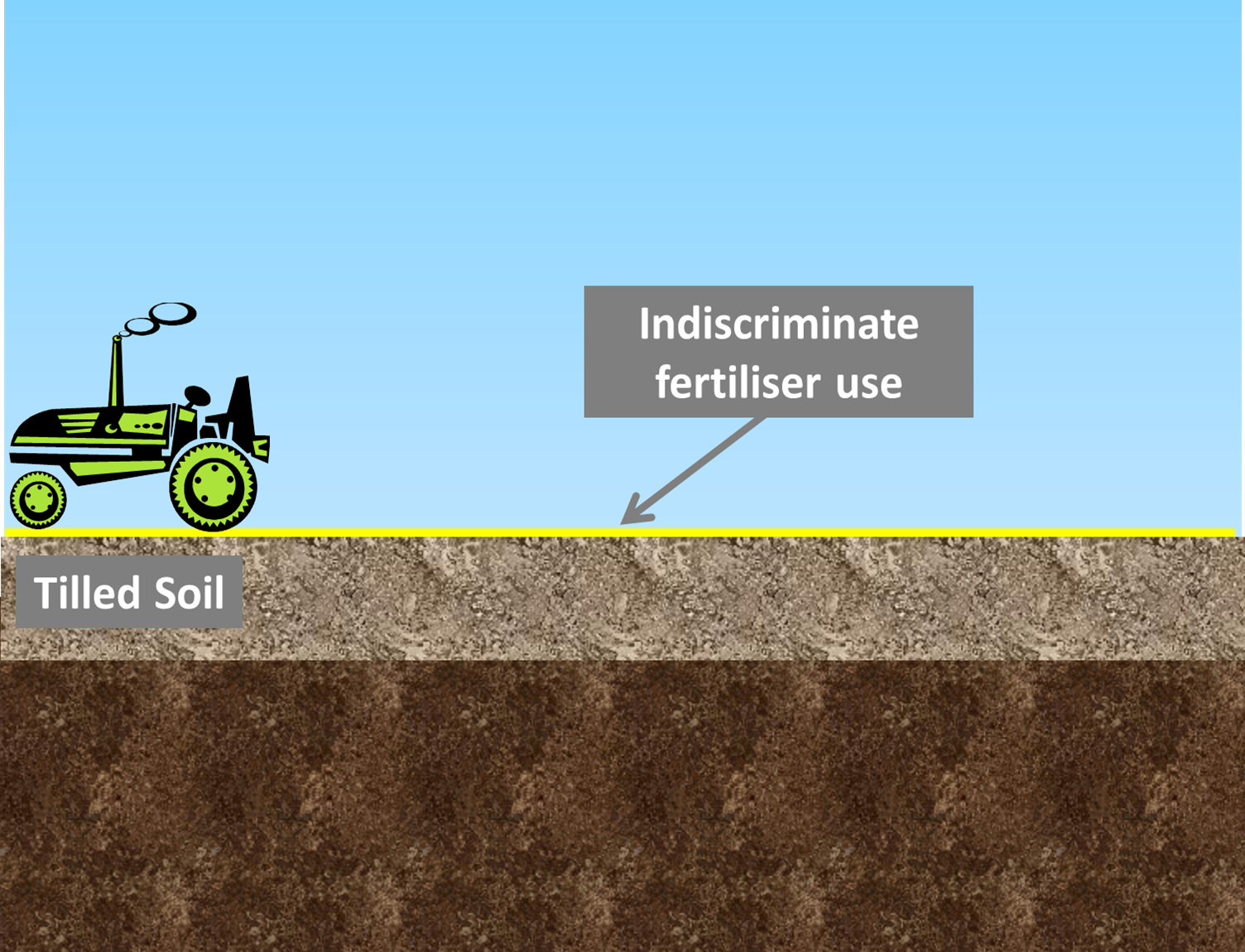
| Next, farmers ‘till’ the soil, replacing the topsoil with deeper, more nutrient-rich soil. Usually farmers will then indiscriminately fertilise the entire field. This is wasteful, as some fertiliser will land on areas which already have abundant nutrient levels. | |
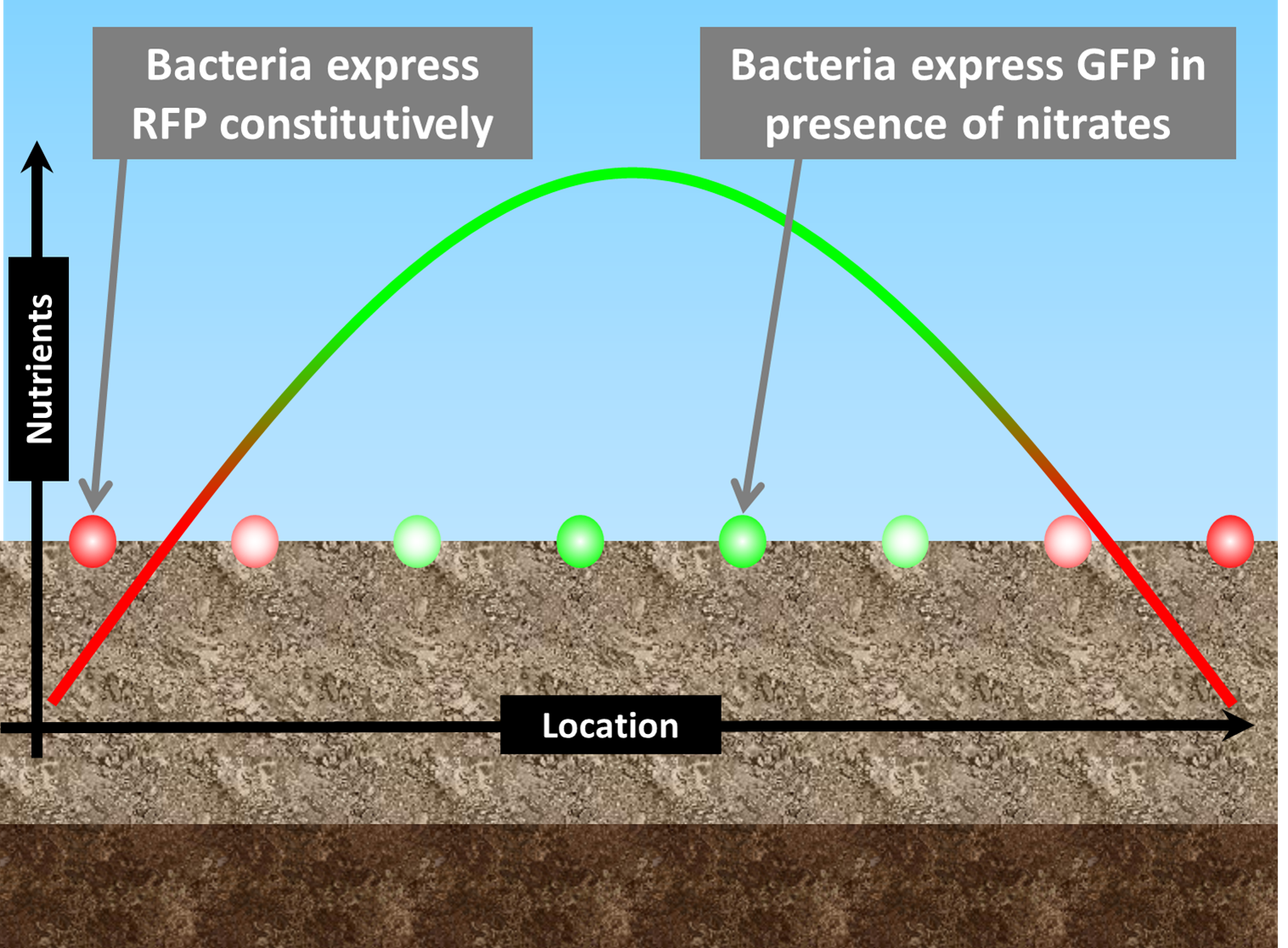
| Instead, we plan to spread agrEcoli bacteria, encapsulated in beads, onto the soil. Since the topsoil has been tilled, it is representative of deeper soil. | |
| agrEcoli bacteria express RFP constitutively, and express GFP in the presence of nitrates. By using a UV light and spectrophotometer attached to a tractor, farmers can infer the nutrient level of soil from the ratio of Red to Green detected. | |
| Using agrEcoli, farmers can map the nutrient content of their fields. This means they can fertilise only the areas that need it, saving up to £45 per hectare http://www.soyl.co.uk/Default.aspx 1. |
References
| http://www.soyl.co.uk/Default.aspx 1 | SoylSense Home page |
 "
"