Team:Imperial College London/Lab Diaries/XylE team
From 2010.igem.org
| Line 50: | Line 50: | ||
<td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:top;"> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>mini-prep kit of XylE-transformed E.coli (already overnight grown)</li> | <li>mini-prep kit of XylE-transformed E.coli (already overnight grown)</li> | ||
| Line 56: | Line 56: | ||
</td> | </td> | ||
</tr> | </tr> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<tr> | <tr> | ||
<td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
| Line 79: | Line 65: | ||
<td style="background-color:#eeeeee;height:100px;width:100px;text-align:top;"> | <td style="background-color:#eeeeee;height:100px;width:100px;text-align:top;"> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Starting of “Testing expression of XylE in E.coli” objective</li> | <li>Starting of “Testing expression of XylE in E.coli” objective</li> | ||
| Line 85: | Line 71: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>gel analysis of mini-prep derived XylE plasmid.(requires first digestion of the vector with restriction enzymes)</li> | <li>gel analysis of mini-prep derived XylE plasmid.(requires first digestion of the vector with restriction enzymes)</li> | ||
| Line 135: | Line 121: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>midi-prep XylE E.coli (2hrs)</li> | <li>midi-prep XylE E.coli (2hrs)</li> | ||
| Line 141: | Line 127: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>gel extraction kit on XylE gene trapped in agarose </li> | <li>gel extraction kit on XylE gene trapped in agarose </li> | ||
| Line 147: | Line 133: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>3A assemply: make replica plates (overnight)</li> | <li>3A assemply: make replica plates (overnight)</li> | ||
| Line 153: | Line 139: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Mini-prep XylE E.coli</li> | <li>Mini-prep XylE E.coli</li> | ||
| Line 159: | Line 145: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Midi-prep XylE E.coli</li> | <li>Midi-prep XylE E.coli</li> | ||
| Line 165: | Line 151: | ||
</td> | </td> | ||
</tr> | </tr> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<tr> | <tr> | ||
<td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | <td style="background-color:#FFCC66;width:100px;text-align:top;"><b>Afternoon</b> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>restriction digestion of XylE for 3A assemply</li> | <li>restriction digestion of XylE for 3A assemply</li> | ||
| Line 193: | Line 162: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>3A assemply of vector, XylE and J23101 promoter </li> | <li>3A assemply of vector, XylE and J23101 promoter </li> | ||
| Line 200: | Line 169: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>*primers arrived!</li> | <li>*primers arrived!</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>PCR extension of XylE and GFP, round 1</li> | <li>PCR extension of XylE and GFP, round 1</li> | ||
| Line 211: | Line 180: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 278: | Line 247: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>PCR extention of His-GFP-flag round 2</li> | <li>PCR extention of His-GFP-flag round 2</li> | ||
| Line 284: | Line 253: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>midi-prep registry obtained J23101</li> | <li>midi-prep registry obtained J23101</li> | ||
| Line 291: | Line 260: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>gel analysis of overnight ligation and gel separation</li> | <li>gel analysis of overnight ligation and gel separation</li> | ||
| Line 297: | Line 266: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>make replica plate and catechol assay plate </li> | <li>make replica plate and catechol assay plate </li> | ||
| Line 303: | Line 272: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>catechol assay</li> | <li>catechol assay</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
| Line 330: | Line 283: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel purification of linker-XylE-Spe PCR construct</li> | <li>Gel purification of linker-XylE-Spe PCR construct</li> | ||
| Line 337: | Line 290: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>restriction digestion, gel analysis and gel purification of registry obtained J23101 </li> | <li>restriction digestion, gel analysis and gel purification of registry obtained J23101 </li> | ||
| Line 345: | Line 298: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>gel purification of overnight ligation</li> | <li>gel purification of overnight ligation</li> | ||
| Line 352: | Line 305: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>PCR construction of fusion protein</li> | <li>PCR construction of fusion protein</li> | ||
| Line 358: | Line 311: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel analysis and gel purification of fusion Xyl E protein</li> | <li>Gel analysis and gel purification of fusion Xyl E protein</li> | ||
| Line 435: | Line 388: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>mini-prep of promoter-XylE transformed E.col</li> | <li>mini-prep of promoter-XylE transformed E.col</li> | ||
| Line 442: | Line 395: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>midi-prep of promoter-XylE transformed E.coli</li> | <li>midi-prep of promoter-XylE transformed E.coli</li> | ||
| Line 450: | Line 403: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel purification kit of cut promoter-XylE DNA</li> | <li>Gel purification kit of cut promoter-XylE DNA</li> | ||
| Line 457: | Line 410: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>find optimun wavelength for catechol assays</li> | <li>find optimun wavelength for catechol assays</li> | ||
| Line 463: | Line 416: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>find optimun cell density and catechol concentration for assay</li> | <li>find optimun cell density and catechol concentration for assay</li> | ||
| Line 469: | Line 422: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
| Line 491: | Line 428: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>gel analysis of mini-prepped promoter-XylE</li> | <li>gel analysis of mini-prepped promoter-XylE</li> | ||
| Line 498: | Line 435: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>PCR purification of cut pSB1C3+terminator vector DNA </li> | <li>PCR purification of cut pSB1C3+terminator vector DNA </li> | ||
| Line 505: | Line 442: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>dephosphorylation of the vector+terminator</li> | <li>dephosphorylation of the vector+terminator</li> | ||
| Line 511: | Line 448: | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>GFP 2 PCR with new reverse primer (adopted TEV sequence)</li> | <li>GFP 2 PCR with new reverse primer (adopted TEV sequence)</li> | ||
| Line 576: | Line 513: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>PCR extension of Pveg promoter </li> | <li>PCR extension of Pveg promoter </li> | ||
| Line 582: | Line 519: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel purification of extended Pveg promoter</li> | <li>Gel purification of extended Pveg promoter</li> | ||
| Line 589: | Line 526: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>vector with insert tranformed into E.coli and plate to select for successful transformation</li> | <li>vector with insert tranformed into E.coli and plate to select for successful transformation</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>prepare overnight cultures of pVeg-RBS transformed E.coli </li> | <li>prepare overnight cultures of pVeg-RBS transformed E.coli </li> | ||
| Line 601: | Line 538: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>midi-prep of Pveg-RBS vector</li> | <li>midi-prep of Pveg-RBS vector</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
| Line 628: | Line 549: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel analysis and purification of the GFP-TEV construct</li> | <li>Gel analysis and purification of the GFP-TEV construct</li> | ||
| Line 634: | Line 555: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>restriction digestion and gel purification of thr extended Pveg promoter</li> | <li>restriction digestion and gel purification of thr extended Pveg promoter</li> | ||
| Line 642: | Line 563: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Amplification PCR for XylE-2</li> | <li>Amplification PCR for XylE-2</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel-purification of amplification-PCR for XylE-2</li> | <li>Gel-purification of amplification-PCR for XylE-2</li> | ||
| Line 653: | Line 574: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
</td> | </td> | ||
| Line 732: | Line 653: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel-analysis of amplification-PCR for XylE-2</li> | <li>Gel-analysis of amplification-PCR for XylE-2</li> | ||
| Line 738: | Line 659: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>midi-prep of Pveg-RBS vector</li> | <li>midi-prep of Pveg-RBS vector</li> | ||
| Line 744: | Line 665: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>In preparation of ''catechol cell-death assay'' tranformation of Top10 cells with CMR plasmid (psb1C3)</li> | <li>In preparation of ''catechol cell-death assay'' tranformation of Top10 cells with CMR plasmid (psb1C3)</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
| Line 780: | Line 685: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>Gel-purification of amplification-PCR for XylE-2</li> | <li>Gel-purification of amplification-PCR for XylE-2</li> | ||
| Line 786: | Line 691: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>plating and overnight incubation of transformed top10 CMR</li> | <li>plating and overnight incubation of transformed top10 CMR</li> | ||
</ul> | </ul> | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
<li>overnight cultures for catechol cell death assay: Top10 cells XylE/CMR </li> | <li>overnight cultures for catechol cell death assay: Top10 cells XylE/CMR </li> | ||
| Line 802: | Line 707: | ||
</td> | </td> | ||
| - | <td style="background-color:#eeeeee;height:100px;width:200px;text-align: | + | <td style="background-color:#eeeeee;height:100px;width:200px;text-align:left;"> |
<ul> | <ul> | ||
</td> | </td> | ||
Revision as of 17:00, 27 October 2010
| Lab Diaries | Overview | Surface Protein Team | XylE Team | Vectors Team | Modelling Team |
| Here are the technical diaries for our project. We've split them up into three lab teams and the modelling team. We think it's really important that absolutely anyone can find out what we've been doing. For a really detailed look at what we did, and when, you've come to the right place! | |
| XylE Team |
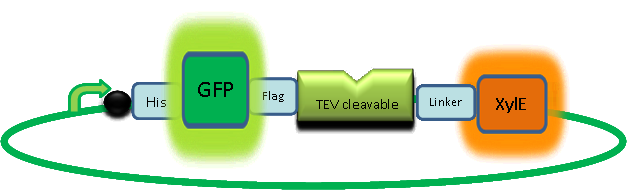
|
Objectives:
|
| Week 6 | ||||||||||||||||||
|
we constructed the standard E.coli promoter J23101 with sticky ends. These ends are complementary to restriction sites made by EcoRI and SpeI enzyme. This promoter will be later used in 3A assemply to construct a promoter-RBS-XylE design in a psB1C3 vector. E.coli will be transformed with this final construct plasmid to assess XylE activity and characterization. It will also be one of the submitted biobricks.
these cultures are going to be used tomorrow for mini-prepping. Miniprep will allow us to isolate E.coli's plasmid DNA(which contains the XylE gene). Friday, 13-Aug-2010
Mini-prep is usually used to confirm that our gene of interest has not been changed in any way, as the isolated plasnid id sent for sequencing. However, since XylE was taken from the registry, we assume that it is fine and no sequencing is required. The mini-prep will later be used for the midi-prep (that gives out higher yeilds of DNA needed for cloning).
|
| Week 7 | ||||||||||||||||||
|
Tuesday, 17-Aug-2010
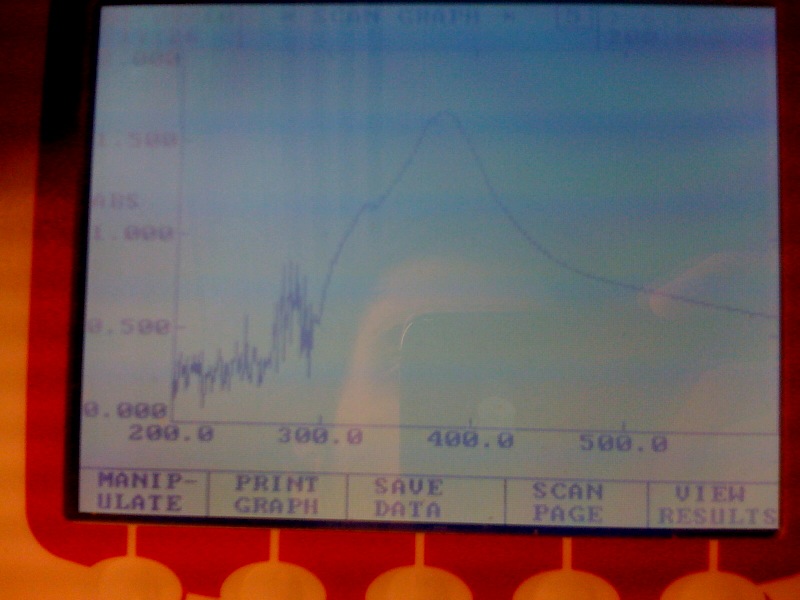
gel analysis of XylE, J23101 promoter and pSB1C3 vector samples to determine the volume ratios of samples to be used for 3A assemply ligation
Thursday, 19-Aug-2010
Friday, 20-Aug-2010 The J23101 gene in a biobrick vector containg RFP gene
|
| Week 8 | ||||||||||||||||||
|
Monday, 23-Aug
Tuesday, 24th-Aug
Wednesday, 25th-Aug Performed gel analysis on the purified XylE and J23101 to obtain ratios for ligation. First gel was scrapped as it produced appauling(explanation for Nick:really bad) results, 2nd gel run was successful.
Thursday, 26th-Aug
Friday, 27th-Aug
Saturday, 28th-Aug
Sunday, 29th-Aug
|
| Week 10 | ||||||||||||||||||
|
Tuesday 7th Sept
Wednesday, 8th Sep
Thursday, 9th Sep
Friday, 10 sep The transformation was a SUCCESS. 2x replica plates were made plate 1# 1-6 plate #2 6-11; colony 6 and colony 9 of plates 1 and 2 respectively were transfered into a 5ml liquid culture + 5ul CmR. These will later be turned into glycerol stocks. After the replica plates have grown up mini preps on a number of colonies shall be performed - this hopefully will eliminate the contaminating plasmid DNA. This will be followed by a midi prep. Saturday, 11 Sep The transformation of E.Coli with PSB1-C3 with insert did not work :( |
| Week 11 | ||||||||||||||||||
|
Monday 13th september Overnights weren't set up on sunday so they were made up alongside some assay cultures. J23101-XylE-B0014 colonies 8 and 10 of the replica plate #2 were picked. Chris also provided a replica plate containing 3k3 vector colonies. This was over a year old and he was unsure whether it was the correct plasmid or if the cells would grow up. I picked all available colonies 102 150 151 and 260 of kanamycin resistance. Lastly for assays 2x LB 2xM9 cultures were made 5ml +5ul antibiotic. Tues 14th September
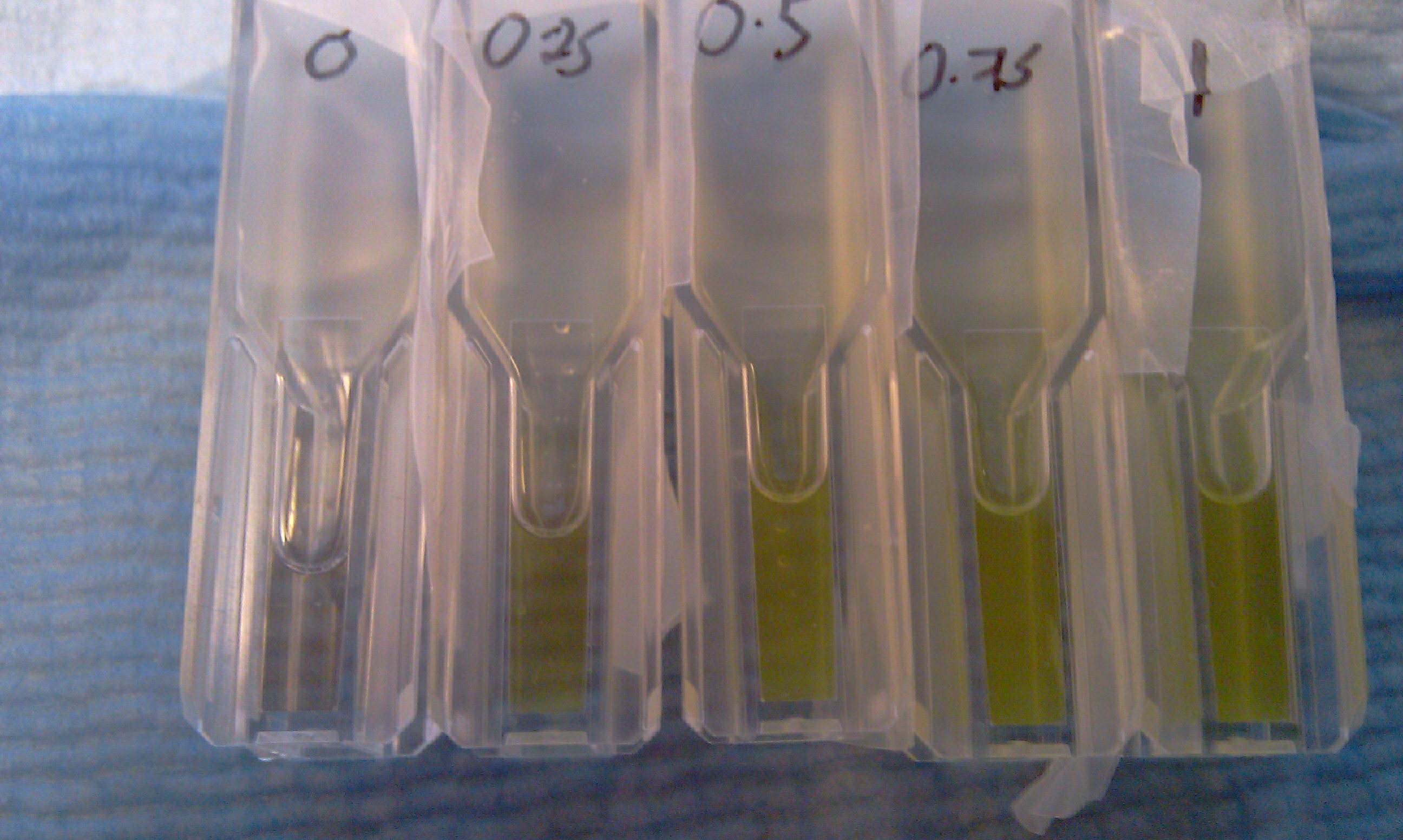
The assay was carried out with E.coli, top ten spcies, transformed with J23101-XylE-B0014 in pSB1C3 vector.The overnight culture wastransfered in new medium this morning for 4 hrs before assaying. LB medium was used for dilutions and blank. Catechol was diluted in ddH2O. Data analysis of the assay
Piotr:
Weds 15th September
Thurs 16th September
- digest with XbaI and SpeI - PCR reactions with the following primers added: HIS-GFP, XylE-Xba and the other sample with HIS-GFP and GFP-flag The PCR reactions were not very conclusive on the gels but digests allowed to determine that colonies 1->5 seem to have the right sizes of DNA in them and on Friday they will be prepared to be sent off for sequencing
Friday 17th September
The gel purifications of 3k3 vector and XylE were used for ligation. 3K3 was dephosphorylated and then ligated in a ratio 5:1 with the insert. This was left overnight and Chris tried to transform with it on sunday.. ligation and transformation failed :-( |
| Week 12 |
|
Monday 20th Sept Performed ligation again. Errors in the sequencing we received back regarding J23101-XylE-B0014. One was a log error (in XylE) phew. The other is in PSB1C3 out of the scar site and should be OK. Chris is currently assembling the pVEG promoter+ RBS in a vector. Once he has finished this, I will combine XylE-GFP and XylE with this promoter for comparison characterization with J23101 (in E.coli and Bacillus) Tues 21st Sept
Wed 22nd Sept
Thurs 23rd Sept First day of AUTUMN!!
Fri 24th Sept
|
| Week 13 |
|
Monday 27th Sept
Tues 28th Sept
Weds 29th Sept
Thurs 30th Sept
Friday 1st Oct
ahahahaha awesome. hey you :)
|
have a look :)it's from John Hoppkins wiki 2008. There are one or two good ones! Love: Before I heard the doctors tell The dangers of a kiss; I had considered kissing you. The nearest thing to bliss. But now I know biology and sit and sigh and moan; six million mad bacteria and I thought we were alone!
|
| Week 14 |
|
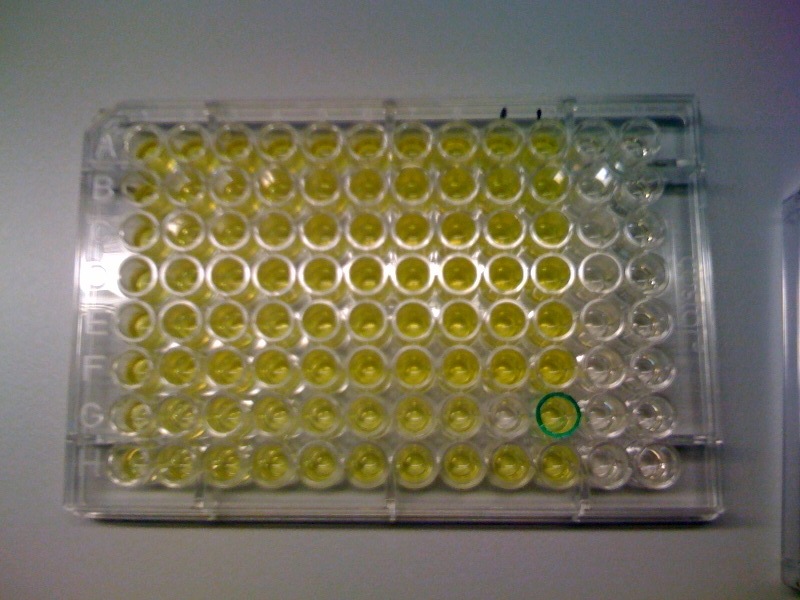
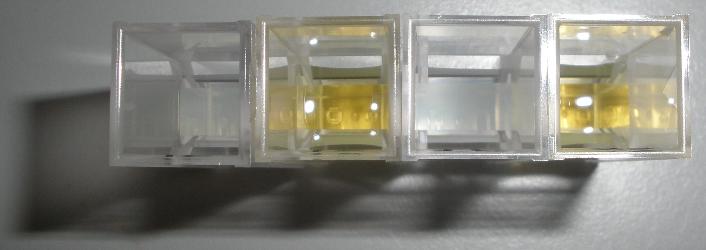
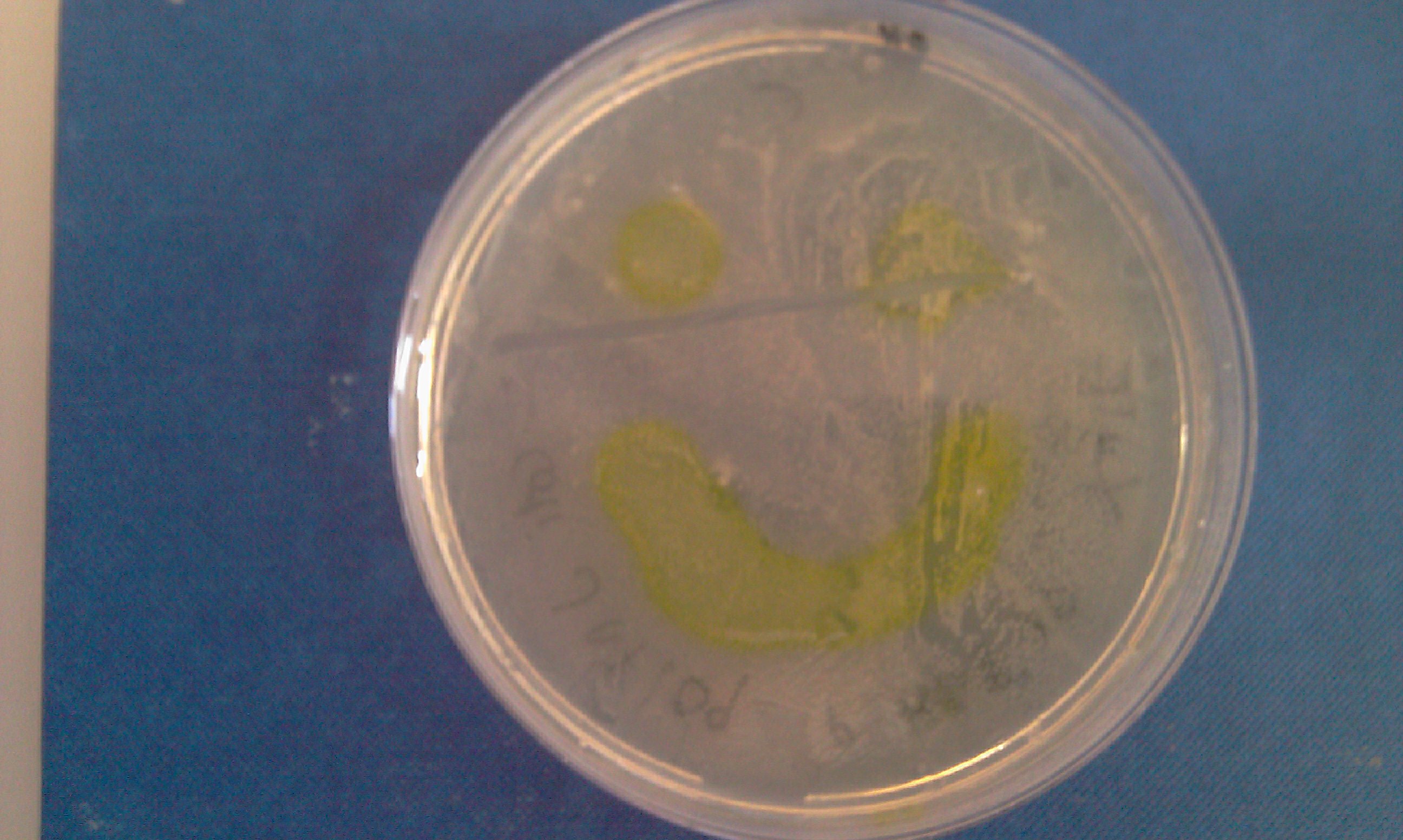
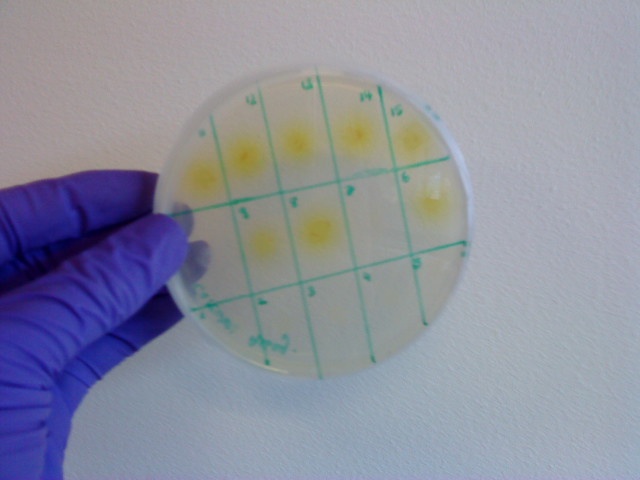
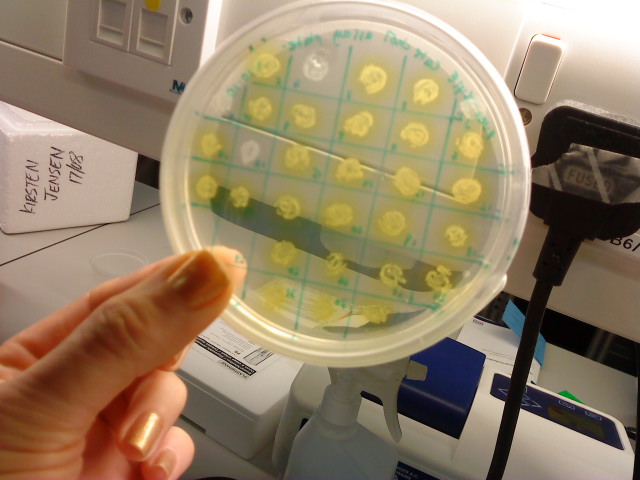
Monday 4th Oct Did a replica plate and catechol assay plate/ minis 2 6 7 9 12 16. (Successful Pveg-XylE transformation) Tues 5th Oct 4 16 colonies were background the rest turned yellow. Mini prepped 2 6 7 9 12 16. Made overnight assay cultures of 3k3-XylE and C3-xylE Weds 6th Oct
Friday 8th Oct
Sunday 10th of the 10th of the 2010!!!!! and im in LAB
|
| Week 15 |
|
Monday 11th Oct
Tues 12th Oct
Weds 13th Oct mini prepped 3k3-Pveg-XylE-B0014 and went to the school workshop! set off overnight midi of culture 9 Thurs 14th Oct Midi prepped 3k3-Pveg Fri 15th Transformed all my constructs into T0P10 3k3-J23101/pveg and PSB1C3-J23101/PVEG |
 "
"