Team:Tsinghua/project/con
From 2010.igem.org
(New page: __NOTOC__ {{:Team:Tsinghua/header}} <html> <body id="bd_project"> <div id="content_box"> </html> {{:Team:Tsinghua/leftbar}} <html> <script> navlist=new Array("Background", "Project Outlin...)
Newer edit →
Revision as of 16:38, 27 October 2010

Outline
In order to realize the production of antibodies in prokaryotic cells, we firstly investigate the whole process of antibody production in mammalian immune system and try to simulate the whole process via the methods and concepts of synthetic biology. Finally, we successfully constructed the desired system. The comparison between the key steps involved in antibody generation and corresponding synthetic methods are shown in the chart below.
| Antibody Production | E Coli. Production System | ||||
|---|---|---|---|---|---|
| VDJ Recombination | Preparation | RSS Sequence |  |
Landing Pad Insertion | Module I: Recombination System |
| VDJ Recombinase |  |
Helper Plasmid Insertion | |||
| VDJ Library |  |
Donor Plasmid Construction | |||
| Recombination |  |
DP Insertion and Recombination Induction | |||
| Somatic Hypermutation |  |
||||
| CBD-based Microarray | Module II: Selection System | ||||
| Antigen Selection |  |
||||
| ToxR-based Transmembrane Pathway | |||||
| Cooperation With Macquarie_Australia | |||||
Generally speaking, antibody production in our project can be divided into two modules.
Module I: Generation of antibody library

Module II: Selection of specific antibodies

Project Design
Module 1
Antibodies Library Diversity & Randomicity
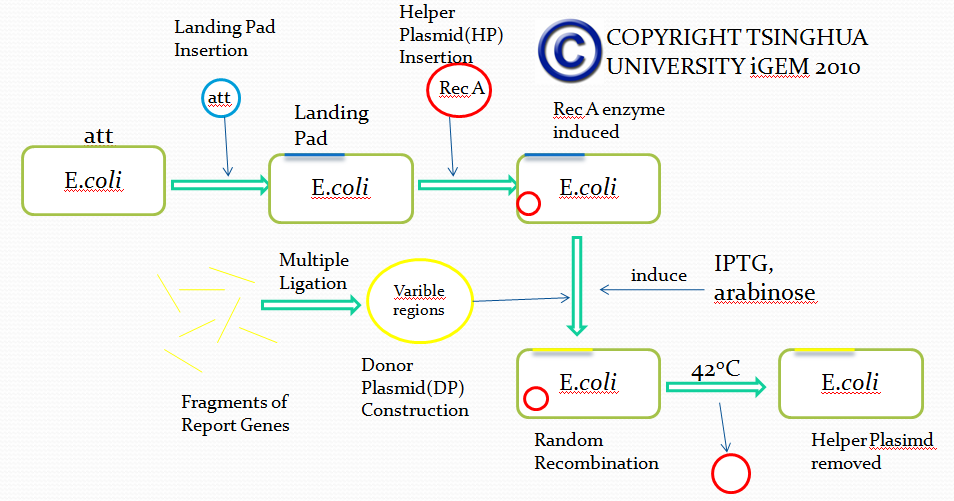
The first module is carried out by a novel method called E coli in vivo recombination system. The theoretical and experimental basis comes from the work of Thomas E. Kuhlman and Edward C. Cox. We managed to modify their system, wishing to utilize it for simulation of antibody recombination.
Firstly, we try to insert a DNA segment called”Landing Pad(LP)” into the specific sites of E coli via Att recombination. Landing pad sequence consists of the following parts(from 5’ to 3’ in the sequence):
1)25bp-long random sequence
2)15bp-long recognition sequence of restriction enzyme I-scel
3) antibiotic resistance gene used for antibody selection
4) 15bp-long recognition sequence of restriction enzyme I-scel(corresponding to 2)
5) 25bp-long random sequence (corresponding to 1)
After integrating DNA segment of Landing pad into the genome of E coli, we completed the genetic engineering of the genome of E coli. All the five parts of landing pad are designed for subsequent recombination.
Besides, in order to achieve different recombination goals, we designed several landing pads of different sequences.
PLEASE refer to 'Landing Pad Construction and Insertion' to learn about the details of Landing Pad design.
PLEASE refer to this 'link' to learn more about ATT recombination.
After the integration of landing pad, we have to insert another DNA fragment called Helper Plasmid(HP), which provides necessary tools for in vivo recombination.
In this plasmid(Helper Plasmid), there are two genes used for recombination.
The first one encodes restriction enzyme I-scel after induction of L-arabinose. I-scel is able to recognize the 15bp-length sequence flanking a target DNA fragment and cut out the fragment, therefore providing DNA segments for recombination mediated by other components.
The second one encodes the enzyme responsible for recombination called Lamda-Red. With the presence of IPTG, Lamda-Red will recombine DNA fragments flanked by the same DNA sequence recognized by Lamda-Red.
In addition, Helper Plasmid contains a specific temperature-sensitive replicon, which is used to control the replication via temperature changes. We can remove the replicon when necessary.
PLEASE refer to 'HP insertion' to learn more about Helper Plasmid.
Up till now, we successfully engineered a ‘recepient strain’. Then we have to transform the ‘recepient strain’ with the Donor Plasmid(DP) containing recombination fragments. Donor Plasmid contains several Insertion Fragments and each Insertion Fragment contains following five parts:
1) one 15bp-long restriction enzyme I-scel recognition site
2) one 25bp-long random sequence
3) one fragment for insertion and recombination
4) one 25bp-long random sequence
5) one 15bp-long restriction enzyme I-scel recognition site(in correspondence with 1)
(Note: the order of the five parts differ from that of landing pad)
As we previously mentioned, Donor plasmid contains several Insertion Fragments. Based on the flanking landing pad sequence, we can ascribe Insertion Fragments to the same group as long as the flanking landing pad sequences of those Insertion Fragments are the same.
In order to achieve different goals, we design different Donor Plasmids, different Donor Plasmids contain different number of Insertion Fragments, which belong to different groups.
PLEASE refer to ‘DP construction’ to learn about the details of Donor Plasmid and Insertion Fragments and the strategies employed in our design.
As we all know, antibody diversity mainly lean on the number of the fragments used for recombination. Therefore, in order to mimic the antibody generation process, we need to construct Donor Plasmid which contains large sum of Insertion Fragments. Theoretically, we can construct Donor Plasmid of this kind. However, due to the time limit of the competition, we didn’t try to construct DP containing many Insertion Fragments.
In spite of this, in order to meet the needs, we designed two methods to construct plasmids on a large scale and attempted the usage in parts of our Donor Plasmids.
PLEASE refer to 'DP construction and Strategies' for the methods of rapid plasmid construction.
Besides, we have applied for two BBFRFC, which are BBFRFC61 and BBFRFC 62 respectively.
Then, after transformation of the prepared ‘recepient strain’ with Donor Plasmid and the induction of corresponding agents, I-scel enzyme cuts out DNA fragments from DP and LP, generating numerous Insertion Fragments. Besides, antibiotic resistance gene integrated into E coli genome is also cut out, providing recombination sites for Insertion Fragments.
Provided with Insertion Fragments mentioned above and Lamda-Red expressed by Donor Plasmid, Insertion Fragments recombine with the corresponding landing pad sequence in the genome and thus get inserted into the correct site of the genome. Based on our design of Landing pad, we can ensure that only one Insertion Fragment from one group is randomly inserted into the genome, therefore mimicking the VDJ recombination process.
PLEASE refer to ‘DP Insertion and Recombination Induction’ to learn more about this part.
Engineered E coli cells which have undergone successful recombination can be selected out of the pool through specific screening methods. After tests on different strains, our project achieve a recombination rate above 50% based on our current techniques.
PLEASE refer to ‘Result’ to learn more about the identification of recombination rate.
Module 2
The goal of the second module in our project is aimed at selection of specific antibodies. Due to the maturity of existing antibody screening technique, our work mainly focuses on imitation of mammalian antibody selection, thus providing possible alternatives for existing methods.
The outline of our ideas is to find mechanisms similar to in vivo antibody selection and take in,to account the industry production costs, controllability and reliability, thus developing new methods for antibody selection. In fact, in mammalian system, antibody selection is achieved through the activation of B lymphocytes and thus the rapid proliferation of the B lymphocytes that express the specific antibodies that bind to antigen. Simply put, the whole process relies on the interaction between antigen epitopes and membrane integral immunoglobins.
To mimic the activation mediated by membrane integral immunoglobins, we develop ToxR-mediated transmembrane activation signal system. In this system, the interaction between antibodies and antigen triggered downstream expression of reporter genes, thus providing signals for selection.
PLEASE refer to ‘ToxR-based Transmembrane Signaling Pathway Method’ for detailed description of this method.
In addition, we develop a technique called ‘Bacterial based Microarray’ for selection purpose. In this method, we combine membrane display technique and high throughput microarray technique, that is, a bacterial based microarray method.
PLEASE refer to ’Bacterial based microarray’ for details.
In order to find methods for antibody selection, we cooperate with other iGEM teams. For example, we talked with Macquarie_Australia iGEM team about the project. The techniques involved in their project might be useful for our selection methods. Therefore, members from both teams worked together to research on this problem.
PLEASE refer to this 'link' to learn more about our cooperation.
Antibody Coding Gene Recombination, also known as V(D)J recombination, somatic recombination, is a mechanism of genetic recombination in the early stages of immunoglobulin (Ig) and T cell receptors (TCR) production of the immune system. V(D)J recombination nearly-randomly combines Variable, Diverse, and Joining gene segments of vertebrates, and because of its randomness in choosing different genes, is able to diversely encode proteins to match antigens.
Our system is aiming at imitating this recombination process, using E.coli as the gene carrier.
It is known to all that regional genes (V, D, J) are flanked by Recombination Signal Sequences (RSSs), and the recombination occurs when VDJ recombinase are expressed. We choose RecA enzyme to induce the recombination, while use I-Sel enzyme to cut the genome, just mimicking the process happened in B cells.
Comparison between natural antibody production and E. Coli system
VDJ Recombination vs Lading Pad Recombination
Antibody Coding Gene Recombination, also known as V(D)J recombination, somatic recombination, is the mechanism by which immunoglobulin (Ig) and T cell receptors (TCR) are generated in the immune system. Through V(D)J recombination, Variable, Diverse, and Joining gene segments are randomly combined, thus encoding various kinds of immunoglobins and T cell receptors to recognize potential antigens. During the process of B cell maturation, gene segments are rearranged and thus transcribe mRNA that can produce diverse types of antibody for recognition of billions of potential epitopes. The rearrangement brings three gene segments (termed V, D, and J) in close proximity, which are then joined together head to tail, the process of which is known as V(D)J recombination based on the V,D,J gene segments involved. Multiple copies of the V, D, and J genes in the human genome are shuffled in the process, and a specific antibody will be generate among millions of other possible combinations. Because gene are joined together permanently, one mature B cell will produce only one specific antibody.
Our system is aimed to mimic this recombination process, using E.coli as the carrier. Through previous research in immunology field, each kind of gene segments (V, D, J) are flanked by Recombination Signal Sequences (RSSs), and recombination occurs when VDJ recombinase recognize the specific signal sequence and mediate the recombination process. In our project, we design a sequence, which carries separated parts of one complete gene and integrate synthesized DNA fragment into the genome of E coli via landing pad method. Then we use I-Sel enzyme to cut the gene segments out of E coli genome, into which gene segments are inserted and choose RecA enzyme to recognize the previously designed signal sequence flanking our target sequence and recombine the gene segments, just like what recombinase accomplish in B cells. The whole process is called landing pad recombination.
Somatic hyper-mutation vs Junctional mutation
Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes. During proliferation, the B cell receptor locus undergoes an extremely high rate of somatic mutation that is at least 105-106 fold greater than the normal rate of mutation across the genome. Variation is mainly in the form of single base substitutions, with insertions and deletions being less common. These mutations occur mostly at “hotspots” in the DNA, known as hypervariable regions. These regions correspond to the complementarity determining regions; the sites involved in antigen recognition on the immunoglobulin. This directed hypermutation allows for the selection of B cells that express immunoglobulin receptors possessing an enhanced ability to recognize and bind a specific foreign antigen.
In addition, the cross ligation, namely Landing pad recombination, has higher opportunity to mutate in the link sequence. The introduction of 4 junctional shuffler 64 will meet the need of the rest 107 diversity, perfectly undertaking the work in the antibody diversity construction that hyper-mutation does in the highly variable region. In this way, we have finished a building library, waiting for the specialized antigen to select.
Antigen-specific Selection vs CBD-Based Microarray and ToxR-Based Transmembrane pathway method
In mammalian immune system, billions of antibodies will be generated. Then how can the immune system find out specific antibody that indiscriminately recognize the specific antigen that comes in. Generally speaking, due to the fact that one mature B cell only generates one specific immunoglobin, once the membrane integral immunoglobin bind to the specific antigen, a series of intracellular response, such as gene activation and cell differetiation will be triggered in B cell. Then, B cells expressing the specific antibodies that recognize the specific antigen will proliferate and generate enormous amount of antibodies that help eliminate the specific antigen.
Simply put, antibody selection is accomplished through the specific interaction between the membrane immunoglobin and the specific antigen. Therefore, in our project, we also plan to mimic this process through interaction between the antibodies generated through landing pad recombination and the antigen we target. CBD-based Microarray and ToxR-based Transmembrane pathway are the two methods by which we achieve this goal. CBD domain, which interacts with cellulose, potentially provides us way to locate our E.coli to the microarray coated with cellulose. OmpA (outer membrane protein A), displays the antibody and antigen out to the membrane surface. Then by applying the antibodies generated through landing pad recombination to the microarray, antibodies that bind to the antigen will be retained and thus selected out of the pool of antibodies.
Alternatively, the ToxR-based transmembrane pathway method shows a different strategy for us to seek the right folded antibody. Once the antigen comes, ToxR outer part will form dimer shape and the downstream signal occur, supplying the approach of reporters, such as GFP, to examine and pick out what we want.
 "
"






 TOP
TOP