Team:Penn State/Project
From 2010.igem.org
(→Results) |
|||
| Line 34: | Line 34: | ||
Imagine a glass jar that is has been sealed off from the outside air. We want to be able to tell how good the jar is at keeping oxygen out using expression of an anaerobic fluorescent protein (AFP) in the bacteria as an indicator. To set up the experiment, the jar is filled with agar populated by E. coli which will consume all the available oxygen in a short period of time. The bacteria contains our plasmid which includes an oxygen-sensing promoter and quorum sensing genes. In the event that oxygen leaks into the jar, the top layer of bacteria react by expressing AFP. | Imagine a glass jar that is has been sealed off from the outside air. We want to be able to tell how good the jar is at keeping oxygen out using expression of an anaerobic fluorescent protein (AFP) in the bacteria as an indicator. To set up the experiment, the jar is filled with agar populated by E. coli which will consume all the available oxygen in a short period of time. The bacteria contains our plasmid which includes an oxygen-sensing promoter and quorum sensing genes. In the event that oxygen leaks into the jar, the top layer of bacteria react by expressing AFP. | ||
| - | [[Image:jar_first.png|600px]] | + | <center>[[Image:jar_first.png|600px]]</center> |
However, this is a weak indicator. If only the top layer of cells is fluorescing, it will be hard to detect a change at all! Quorum sensing solves this problem by sending a signal to all the surrounding bacteria that will cause them to begin producing AFP. | However, this is a weak indicator. If only the top layer of cells is fluorescing, it will be hard to detect a change at all! Quorum sensing solves this problem by sending a signal to all the surrounding bacteria that will cause them to begin producing AFP. | ||
| - | [[Image:jar_before.png|600px]] | + | <center>[[Image:jar_before.png|600px]]</center> |
In this way, only when oxygen-sensing and quorum-sensing are used together will the AFP indicator be a strong enough signal to measure. | In this way, only when oxygen-sensing and quorum-sensing are used together will the AFP indicator be a strong enough signal to measure. | ||
| - | [[Image:jar_final.png|600px]] | + | <center>[[Image:jar_final.png|600px]]</center> |
A reliable modular lysis mechanism through quorum sensing could be used in many different fields. As engineered microorganisms are becoming more widely considered for use in the open environment, concerns are growing about possible detrimental effects these microbes could have on ecosystems if they were ever to grow and multiply past expectations. A quorum-based lysis "safety" mechanism would only require one cell to receive a predetermined signal in order to eliminate the entire rogue population, should it escape the area of its intended use. | A reliable modular lysis mechanism through quorum sensing could be used in many different fields. As engineered microorganisms are becoming more widely considered for use in the open environment, concerns are growing about possible detrimental effects these microbes could have on ecosystems if they were ever to grow and multiply past expectations. A quorum-based lysis "safety" mechanism would only require one cell to receive a predetermined signal in order to eliminate the entire rogue population, should it escape the area of its intended use. | ||
| Line 130: | Line 130: | ||
Here is an overview of our genetic circuit. Below, we will take a look at the function of each individual part in detail. | Here is an overview of our genetic circuit. Below, we will take a look at the function of each individual part in detail. | ||
| - | [[Image:Genetic_Circuit_5.jpg|800px]] | + | <center>[[Image:Genetic_Circuit_5.jpg|800px]]</center> |
Our genetic circuit is designed to initially be in an anoxic state. The goal here was to create a circuit with a very strong OFF during anaerobic conditions, and a very strong ON during aerobic conditions. Anaerobic fluorescent protein (AFP) is used as an indicator. | Our genetic circuit is designed to initially be in an anoxic state. The goal here was to create a circuit with a very strong OFF during anaerobic conditions, and a very strong ON during aerobic conditions. Anaerobic fluorescent protein (AFP) is used as an indicator. | ||
===cI mechanism=== | ===cI mechanism=== | ||
| - | [[Image:CImovie.gif|cI mechanism]] | + | <center>[[Image:CImovie.gif|cI mechanism]]</center> |
The Tet inverter behaves somewhat as a constitutive promoter when no promoter is in front of it. Following the Tet inverter is a cI coding sequence (which codes for the lambda repressor). The lambda repressor binds to the lux promoters in the front of the circuit, which are also lambda repressible. This, coupled with the fact that the conditions are anaerobic, provides a very strong "off" switch that does not allow the expression of the Anaerobic Fluorescent Protein (AFP) to occur. | The Tet inverter behaves somewhat as a constitutive promoter when no promoter is in front of it. Following the Tet inverter is a cI coding sequence (which codes for the lambda repressor). The lambda repressor binds to the lux promoters in the front of the circuit, which are also lambda repressible. This, coupled with the fact that the conditions are anaerobic, provides a very strong "off" switch that does not allow the expression of the Anaerobic Fluorescent Protein (AFP) to occur. | ||
===FNR breaks apart in oxygen=== | ===FNR breaks apart in oxygen=== | ||
| - | [[Image:FNRmovie.gif|FNR breaks apart in Oxygen]] | + | <center>[[Image:FNRmovie.gif|FNR breaks apart in Oxygen]]</center> |
FNR can act as a repressor or inducer depending on its position relative to the RBS. In nature, an activated FNR dimer represses aerobic functions while inducing anaerobic functions. | FNR can act as a repressor or inducer depending on its position relative to the RBS. In nature, an activated FNR dimer represses aerobic functions while inducing anaerobic functions. | ||
| Line 147: | Line 147: | ||
===Ptet is repressed, and the positive feedback loop is switched on=== | ===Ptet is repressed, and the positive feedback loop is switched on=== | ||
| - | [[Image:Tet.gif]] | + | <center>[[Image:Tet.gif]]</center> |
The oxygen promoter is followed by 3 protein coding sequences. Two of them are genes that code for the Tet repressor and LuxI (which produces [N-]acyl-homoserine lactone, or AHL). These two parts serve to strengthen the genetic switch once it has been turned on. The Tet repressor binds to the Tet inverter, causing the expression of the lambda repressor to be switched off. Meanwhile, AHL production resulting from the LuxI gene couples with the constitutively-expressed LuxR in order to activate the Lux promoter at the beginning of the circuit. | The oxygen promoter is followed by 3 protein coding sequences. Two of them are genes that code for the Tet repressor and LuxI (which produces [N-]acyl-homoserine lactone, or AHL). These two parts serve to strengthen the genetic switch once it has been turned on. The Tet repressor binds to the Tet inverter, causing the expression of the lambda repressor to be switched off. Meanwhile, AHL production resulting from the LuxI gene couples with the constitutively-expressed LuxR in order to activate the Lux promoter at the beginning of the circuit. | ||
| Line 154: | Line 154: | ||
===AFP expression ramps up=== | ===AFP expression ramps up=== | ||
| - | [[Image:AFP.gif]] | + | <center>[[Image:AFP.gif]]</center> |
Because both the oxygen promoter and the Lux promoter are turned on as part of the positive feedback loop, AFP is now expressing at a very high rate. This is also bolstered by the fact that that the Super RBS is in front of this protein. | Because both the oxygen promoter and the Lux promoter are turned on as part of the positive feedback loop, AFP is now expressing at a very high rate. This is also bolstered by the fact that that the Super RBS is in front of this protein. | ||
| Line 167: | Line 167: | ||
| - | [[Image:D5.jpg|800px]] | + | <center>[[Image:D5.jpg|800px]]</center> |
The oxygen promoters were characterized using a Tecan infinite M1000 microplate reader. Each sample was grown overnight on plates and transferred to a 96 deep-well plate for overnight growth. After 16 hours the samples were tested in the Tecan, where measurements would be taken for around 12 hours. Sometimes, measurement of growth of three successive plates (serially diluted from the preceding one) was needed before the appropriate steady state could be recorded. | The oxygen promoters were characterized using a Tecan infinite M1000 microplate reader. Each sample was grown overnight on plates and transferred to a 96 deep-well plate for overnight growth. After 16 hours the samples were tested in the Tecan, where measurements would be taken for around 12 hours. Sometimes, measurement of growth of three successive plates (serially diluted from the preceding one) was needed before the appropriate steady state could be recorded. | ||
| Line 177: | Line 177: | ||
All measurements were taken using the Tecan Infinite M 1000 spectrophotometer. The following is a graph of the raw fluorescence values recorded by the Tecan for two oxygen promoters, D5 and J6. Each construct was duplicated in the 96-well plate. One was allowed to grow normally under aerobic conditions, while the other sample was covered in 100uL of mineral oil to create an anaerobic environment. Serial dilution of two plates resulted in 3 sets of data for this experiment, as seen below. | All measurements were taken using the Tecan Infinite M 1000 spectrophotometer. The following is a graph of the raw fluorescence values recorded by the Tecan for two oxygen promoters, D5 and J6. Each construct was duplicated in the 96-well plate. One was allowed to grow normally under aerobic conditions, while the other sample was covered in 100uL of mineral oil to create an anaerobic environment. Serial dilution of two plates resulted in 3 sets of data for this experiment, as seen below. | ||
| - | [[Image:Rawoxygen.png|800px]] | + | <center>[[Image:Rawoxygen.png|800px]]</center> |
The next graph shown is the Raw data for OD for the same constructs shown above. The OD is proportional to the number of cells growing in the media, so dividing the fluorescence by these values is approximately proportional to the fluorescence per cell. | The next graph shown is the Raw data for OD for the same constructs shown above. The OD is proportional to the number of cells growing in the media, so dividing the fluorescence by these values is approximately proportional to the fluorescence per cell. | ||
| - | [[Image:RawODoxygen.png|800px]] | + | <center>[[Image:RawODoxygen.png|800px]]</center> |
| - | [[Image:FLUORoverODoxygen.png|800px]] | + | <center>[[Image:FLUORoverODoxygen.png|800px]]</center> |
| - | [[Image:Poxygen.png|800px]] | + | <center>[[Image:Poxygen.png|800px]]</center> |
| - | [[Image:D5promoter.png|800px]] | + | <center>[[Image:D5promoter.png|800px]]</center> |
| - | [[Image:comparison.png|800px]] | + | <center>[[Image:comparison.png|800px]]</center> |
| Line 196: | Line 196: | ||
When designing our project, we were thinking about using the violacein pigment, BBa_K274002, in our genetic circuit. This part is supposed to be just the coding sequence without a promoter. As can be seen by the picture below, the coding sequence appears to have some sort of constitutive promoter attached to it. | When designing our project, we were thinking about using the violacein pigment, BBa_K274002, in our genetic circuit. This part is supposed to be just the coding sequence without a promoter. As can be seen by the picture below, the coding sequence appears to have some sort of constitutive promoter attached to it. | ||
| - | [[Image:Violacein.jpg|400px]] | + | <center>[[Image:Violacein.jpg|400px]]</center> |
| - | [[Image:1107_108.png|800px]] | + | <center>[[Image:1107_108.png|800px]]</center> |
| - | [[Image:1107_116_tetR.png|800px]] | + | <center>[[Image:1107_116_tetR.png|800px]]</center> |
| - | [[Image:146_108_tetR.png|800px]] | + | <center>[[Image:146_108_tetR.png|800px]]</center> |
| - | [[Image:146_116_tetR.png|800px]] | + | <center>[[Image:146_116_tetR.png|800px]]</center> |
Revision as of 15:57, 27 October 2010
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Notebook | Human Practices | Safety | Sponsors |
|---|
Contents |
Overall project
The Penn State Team will be researching and experimenting two different ideas. The first area of study will be creating an oxygen-sensing promoter. This will be done through researching the way that FNR and arcA regulate aerobic/anaerobic respiration in E. coli; engineering applications will be applied to these processes. The second topic we are working on is Bacterial Fireworks, an incorporation of quorum-sensing and oxygen-sensing. Here is a simple example of a problem that our research with oxygen promoters and quorum sensing could solve:
Imagine a glass jar that is has been sealed off from the outside air. We want to be able to tell how good the jar is at keeping oxygen out using expression of an anaerobic fluorescent protein (AFP) in the bacteria as an indicator. To set up the experiment, the jar is filled with agar populated by E. coli which will consume all the available oxygen in a short period of time. The bacteria contains our plasmid which includes an oxygen-sensing promoter and quorum sensing genes. In the event that oxygen leaks into the jar, the top layer of bacteria react by expressing AFP.

However, this is a weak indicator. If only the top layer of cells is fluorescing, it will be hard to detect a change at all! Quorum sensing solves this problem by sending a signal to all the surrounding bacteria that will cause them to begin producing AFP.

In this way, only when oxygen-sensing and quorum-sensing are used together will the AFP indicator be a strong enough signal to measure.

A reliable modular lysis mechanism through quorum sensing could be used in many different fields. As engineered microorganisms are becoming more widely considered for use in the open environment, concerns are growing about possible detrimental effects these microbes could have on ecosystems if they were ever to grow and multiply past expectations. A quorum-based lysis "safety" mechanism would only require one cell to receive a predetermined signal in order to eliminate the entire rogue population, should it escape the area of its intended use.
Project Details
Genetic Circuit
The Penn State iGEM team began work and research in mid May. After learning lab techniques from the grad students, we began researching possible topics. We decided to create a cell lysis device.
Initially, we transformed Lux Promoters, Constitutive Promoters, LuxI + RBS, LuxR + RBS, Lambda Phage lysis devices, Terminators, LacI, LuxR, pLac, CI, RBS, Pigments, FP, LacI + RBS, and CI + RBS.
| Lux Promoters | Lux Promoters | Constitutive Promoters | LuxI + RBS | LuxR + RBS | Lambda Phage Lysis Device | Terminator | AFP |
|---|---|---|---|---|---|---|---|
| [http://partsregistry.org/Part:BBa_K091107 K091107] | [http://partsregistry.org/Part:BBa_R1062 R1062] | [http://partsregistry.org/Part:BBa_J23108 J23108] | [http://partsregistry.org/Part:BBa_C0261 C0261] | [http://partsregistry.org/Part:BBa_J37033 J37033] | [http://partsregistry.org/Part:BBa_K112022 K112022] | [http://partsregistry.org/Part:BBa_B0010 B0010] | [http://partsregistry.org/Part:BBa_K376004 K376004] |
| [http://partsregistry.org/Part:BBa_K091146 K091146] | [http://partsregistry.org/Part:BBa_R0063 R0063] | [http://partsregistry.org/Part:BBa_J23116 J23116] | [http://partsregistry.org/Part:BBa_S01414 S01414] | [http://partsregistry.org/Part:BBa_K112808 K112808] | [http://partsregistry.org/Part:BBa_B1002 B1002] | ||
| [http://partsregistry.org/Part:BBa_J23100 J23100] | [http://partsregistry.org/Part:BBa_B1006 B1006] |
| LuxR | LacI | pLac | cI | RBS | Pigment | FP | LacI + RBS | cI + RBS |
|---|---|---|---|---|---|---|---|---|
| [http://partsregistry.org/Part:BBa_C0062 C0062] | [http://partsregistry.org/Part:BBa_C0012 C0012] | [http://partsregistry.org/Part:BBa_R0010 R0010] | [http://partsregistry.org/Part:BBa_C0051 C0051] | [http://partsregistry.org/Part:BBa_B0033 B0033] | [http://partsregistry.org/Part:BBa_K274100 K274100] | RFP + RBS | [http://partsregistry.org/Part:BBa_J24679 J24679] | [http://partsregistry.org/Part:BBa_K081007 K081007] |
| [http://partsregistry.org/Part:BBa_B0034 B0034] | [http://partsregistry.org/Part:BBa_K274004 K274004] | [http://partsregistry.org/Part:BBa_P0451 P0451] |
Here is an overview of our genetic circuit. Below, we will take a look at the function of each individual part in detail.

Our genetic circuit is designed to initially be in an anoxic state. The goal here was to create a circuit with a very strong OFF during anaerobic conditions, and a very strong ON during aerobic conditions. Anaerobic fluorescent protein (AFP) is used as an indicator.
cI mechanism

The Tet inverter behaves somewhat as a constitutive promoter when no promoter is in front of it. Following the Tet inverter is a cI coding sequence (which codes for the lambda repressor). The lambda repressor binds to the lux promoters in the front of the circuit, which are also lambda repressible. This, coupled with the fact that the conditions are anaerobic, provides a very strong "off" switch that does not allow the expression of the Anaerobic Fluorescent Protein (AFP) to occur.
FNR breaks apart in oxygen

FNR can act as a repressor or inducer depending on its position relative to the RBS. In nature, an activated FNR dimer represses aerobic functions while inducing anaerobic functions.
In the event that conditions change to aerobic in our circuit, oxygen molecules break up the dimer formed by FNR, causing the FNR dimer to detach from the oxygen promoter. Assuming that the oxygen promoter used in this assembly is a repressor, the end result will be that the coding sequences following the promoter will begin to be expressed.
Ptet is repressed, and the positive feedback loop is switched on

The oxygen promoter is followed by 3 protein coding sequences. Two of them are genes that code for the Tet repressor and LuxI (which produces [N-]acyl-homoserine lactone, or AHL). These two parts serve to strengthen the genetic switch once it has been turned on. The Tet repressor binds to the Tet inverter, causing the expression of the lambda repressor to be switched off. Meanwhile, AHL production resulting from the LuxI gene couples with the constitutively-expressed LuxR in order to activate the Lux promoter at the beginning of the circuit.
In addition, the Anaerobic Fluorescent Protein begins to be expressed at a high rate.
AFP expression ramps up

Because both the oxygen promoter and the Lux promoter are turned on as part of the positive feedback loop, AFP is now expressing at a very high rate. This is also bolstered by the fact that that the Super RBS is in front of this protein.
The Experiments
Oxygen Sensor
The design of the oxygen-sensing promoter was based on the dcuC gene of E. coli. The two binding sites for FNR surrounded the ribosome binding site and were systematically shifted upstream and downstream from this site in multiple constructs, causing overlap of the ribosome binding site in some cases. Five constructs were designed using the constitutive promoter J23113 as the basis for the rest of the promoter following the downstream FNR binding site. The construct out of these five that was fully characterized was J6, and its sequence is as follows:

The oxygen promoters were characterized using a Tecan infinite M1000 microplate reader. Each sample was grown overnight on plates and transferred to a 96 deep-well plate for overnight growth. After 16 hours the samples were tested in the Tecan, where measurements would be taken for around 12 hours. Sometimes, measurement of growth of three successive plates (serially diluted from the preceding one) was needed before the appropriate steady state could be recorded.
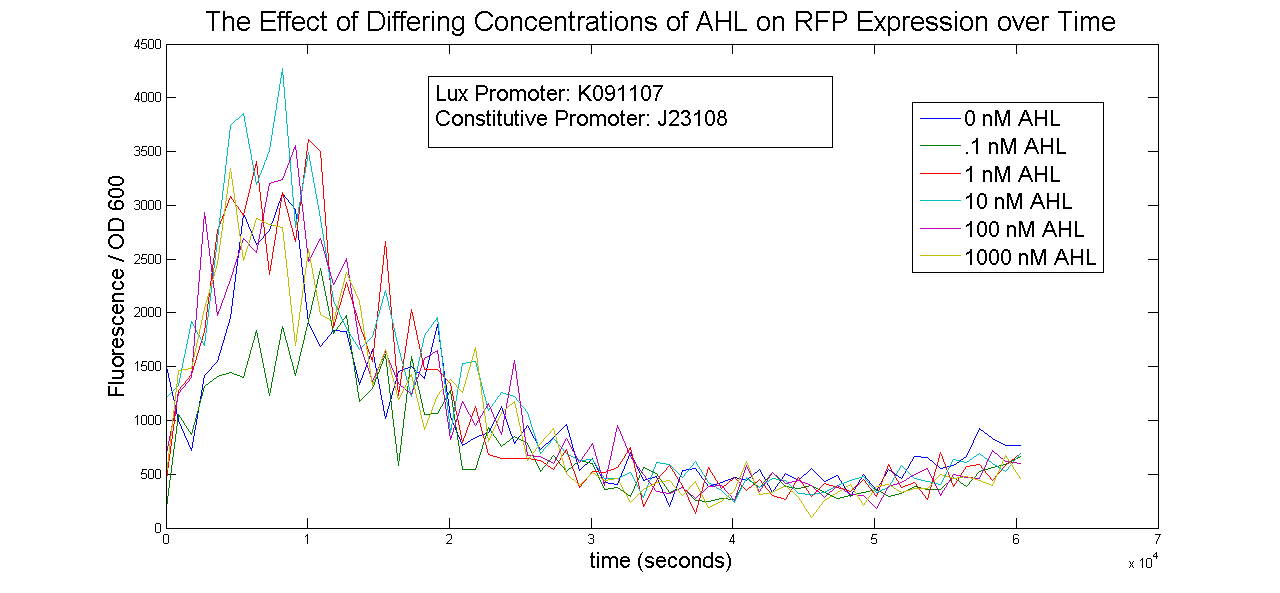
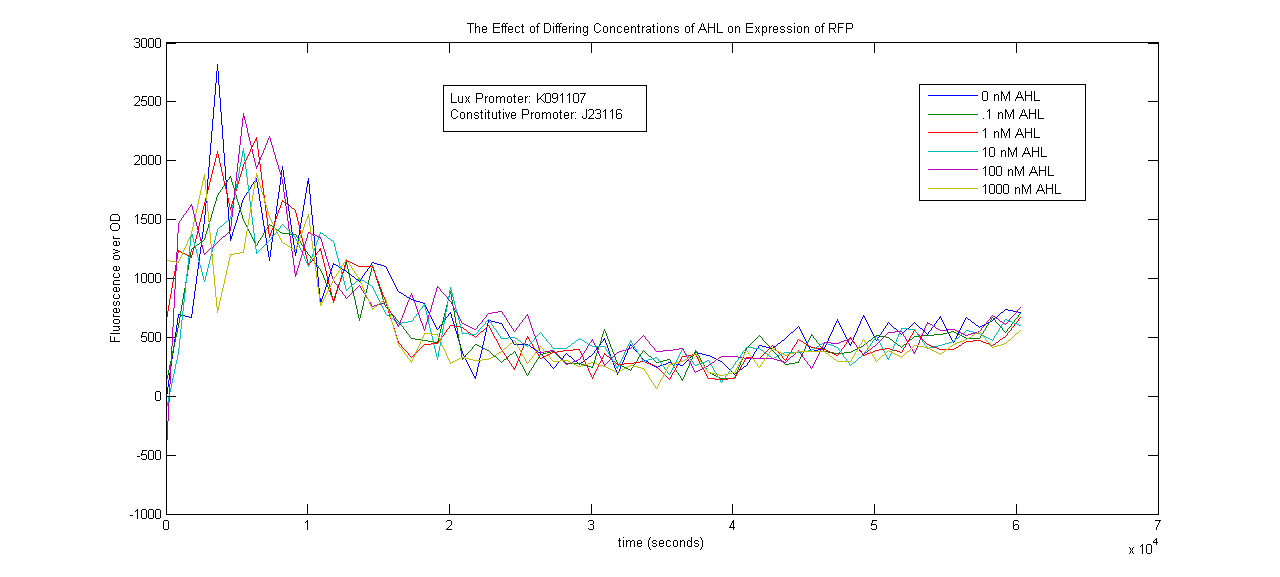
Lux System
Results
All measurements were taken using the Tecan Infinite M 1000 spectrophotometer. The following is a graph of the raw fluorescence values recorded by the Tecan for two oxygen promoters, D5 and J6. Each construct was duplicated in the 96-well plate. One was allowed to grow normally under aerobic conditions, while the other sample was covered in 100uL of mineral oil to create an anaerobic environment. Serial dilution of two plates resulted in 3 sets of data for this experiment, as seen below.

The next graph shown is the Raw data for OD for the same constructs shown above. The OD is proportional to the number of cells growing in the media, so dividing the fluorescence by these values is approximately proportional to the fluorescence per cell.





Characterization of Old BioBrick parts
When designing our project, we were thinking about using the violacein pigment, BBa_K274002, in our genetic circuit. This part is supposed to be just the coding sequence without a promoter. As can be seen by the picture below, the coding sequence appears to have some sort of constitutive promoter attached to it.



 "
"