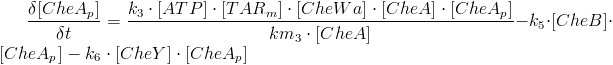Team:SDU-Denmark/project-bc
From 2010.igem.org
(Difference between revisions)
(→Biochemical Modelling) |
(→Biochemical Modelling) |
||
| Line 14: | Line 14: | ||
=== The real system === | === The real system === | ||
Several chemosensory systems exist in bacteria. The one most studied is the chemotaxis of ''E. coli''. Polar clusters of membrane-spanning methyl accepting chemotaxis proteins (MCPs) are positioned in the ends of the rod shaped ''E. coli''.<br> | Several chemosensory systems exist in bacteria. The one most studied is the chemotaxis of ''E. coli''. Polar clusters of membrane-spanning methyl accepting chemotaxis proteins (MCPs) are positioned in the ends of the rod shaped ''E. coli''.<br> | ||
| - | Tar (responding to aspartate), Tsr (responding to serine), Tap (responding todipetides), Trg (responding to galactose) ang Aer (responding to oxygen) are the most common of these receptors seen in ''E. coli'' strains[[https://2010.igem.org/Team:SDU-Denmark/project-bc#References 1]]. Transduction of the signal between the MCPs and the flagella motor is managed by a particular fine-tuned signalling cascade, which is governed by several intracellular proteins.<br | + | Tar (responding to aspartate), Tsr (responding to serine), Tap (responding todipetides), Trg (responding to galactose) ang Aer (responding to oxygen) are the most common of these receptors seen in ''E. coli'' strains[[https://2010.igem.org/Team:SDU-Denmark/project-bc#References 1]]. Transduction of the signal between the MCPs and the flagella motor is managed by a particular fine-tuned signalling cascade, which is governed by several intracellular proteins.<br> |
[[Image: Team-SDU-Denmark-Chemotaxis.png |thumb|center|500px| '''Figure 1:''' Schematics of the bacterial chemotaxis pathways]] | [[Image: Team-SDU-Denmark-Chemotaxis.png |thumb|center|500px| '''Figure 1:''' Schematics of the bacterial chemotaxis pathways]] | ||
<p style="text-align: justify;">Chemotaxis consists of three important phases: reaction, adaptation and relaxation. In the reaction phase, one of the MCPs sense the specific chemical gradient it responds to and the membrane protein activates the CheW enzyme. The active CheW enzyme suppresses the auto-phosphorylation of the CheA enzyme and when no or very little CheA enzyme is phosphorylated, no phosphorylation of the CheY and CheB enzymes occurs. In the end, this increases run, because phosphorylated CheY binds to the flagella motors and induce tumbling. When the bacteria have reached the area with a higher concentration of the chemical in question adaptation sets in.<br><br> | <p style="text-align: justify;">Chemotaxis consists of three important phases: reaction, adaptation and relaxation. In the reaction phase, one of the MCPs sense the specific chemical gradient it responds to and the membrane protein activates the CheW enzyme. The active CheW enzyme suppresses the auto-phosphorylation of the CheA enzyme and when no or very little CheA enzyme is phosphorylated, no phosphorylation of the CheY and CheB enzymes occurs. In the end, this increases run, because phosphorylated CheY binds to the flagella motors and induce tumbling. When the bacteria have reached the area with a higher concentration of the chemical in question adaptation sets in.<br><br> | ||
| Line 27: | Line 27: | ||
The model we will be using to predict biochemical behaviour of the system is a simplification of the real system.<br> | The model we will be using to predict biochemical behaviour of the system is a simplification of the real system.<br> | ||
The simplified model does not consider the normal metabolism in the cell. It only includes the most important enzymes in the normal ''E. coli'' chemotaxis pathway and the proteins responsible for coupling the phototaxis to the chemotaxis.<br> | The simplified model does not consider the normal metabolism in the cell. It only includes the most important enzymes in the normal ''E. coli'' chemotaxis pathway and the proteins responsible for coupling the phototaxis to the chemotaxis.<br> | ||
| - | Below, you will se a schematic representation of the system our biochemical modelling was performed on. The crossed-over enzymes and proteins have not been modelled as a differential equation but considered as a constant in the system.<br | + | Below, you will se a schematic representation of the system our biochemical modelling was performed on. The crossed-over enzymes and proteins have not been modelled as a differential equation but considered as a constant in the system.<br> |
[[Image: Team-SDU-Denmark-biomodelling.png |thumb|400px |center| '''Figure 2''': Schematics of the system we want to make. The enzymes that are crossed-over are considered constant in our system]] | [[Image: Team-SDU-Denmark-biomodelling.png |thumb|400px |center| '''Figure 2''': Schematics of the system we want to make. The enzymes that are crossed-over are considered constant in our system]] | ||
<p style="text-align: justify;">The important enzymes in the system are mathematically represented by a differential equation under the assumption of steady state in the system and modelled in a demo version of the program [http://www.berkeleymadonna.com/ Berkeley Madonna].<br> | <p style="text-align: justify;">The important enzymes in the system are mathematically represented by a differential equation under the assumption of steady state in the system and modelled in a demo version of the program [http://www.berkeleymadonna.com/ Berkeley Madonna].<br> | ||
Revision as of 14:24, 27 October 2010
 "
"