Team:TU Delft/Parts/characterization
From 2010.igem.org
(→Ribosome Binding Site (RBS) sequence characterization) |
|||
| Line 87: | Line 87: | ||
====Ribosome Binding Site (RBS) sequence characterization==== | ====Ribosome Binding Site (RBS) sequence characterization==== | ||
| - | |||
Read more about the [[Team:TU_Delft/Project/rbs-characterization|RBS characterization]]. We used the following 5 constructs to perform the characterization. | Read more about the [[Team:TU_Delft/Project/rbs-characterization|RBS characterization]]. We used the following 5 constructs to perform the characterization. | ||
<partinfo>BBa_K398500 SpecifiedComponents</partinfo> | <partinfo>BBa_K398500 SpecifiedComponents</partinfo> | ||
| Line 97: | Line 96: | ||
Resulting RBS strenghts: | Resulting RBS strenghts: | ||
| + | [[Image:TUDelft_2010_RBS_strength_graph.PNG|thumb|400px|right|Characterization of 5 RBS sequences from the Anderson series]] | ||
{|width="300" | {|width="300" | ||
|<b>Ribosome binding site</b> | |<b>Ribosome binding site</b> | ||
Revision as of 12:26, 27 October 2010
Characterization
Our favorite parts were also the ones best characterized. This page is a quick overview of the results. You can find a complete overview of characterization on the project pages.
Favorite Parts
[http://partsregistry.org/Part:BBa_K398029 BBa_K398029] - ALDH
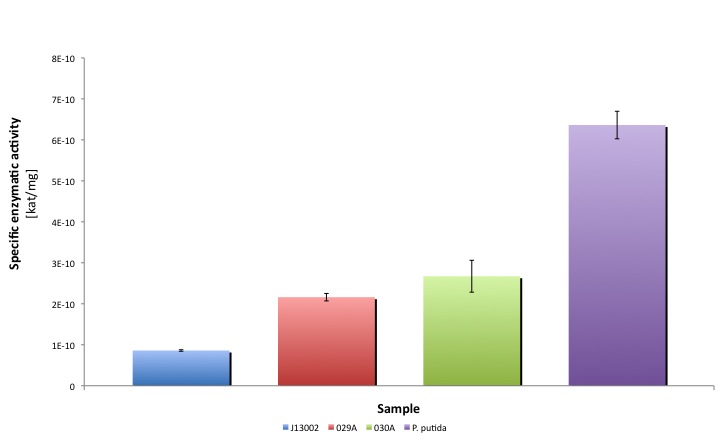
Our results suggest that the recombinant strain E. coli 029A functionally express our biobrick [http://partsregistry.org/Part:BBa_K398029 BBa_K398029]. From the statistical analysis that we performed we concluded that the expression of the biobrick [http://partsregistry.org/Part:BBa_K398006 BBa_K398006] under the promoter-rbs combination [http://partsregistry.org/Part:BBa_J13002 BBa_J23100]-[http://partsregistry.org/Part:BBa_J13002 BBa_J61117] increases the dodecanal dehydrogenase activity in E. coli cell extracts 2-fold. Moreover, the enzymatic activities measured for the construct [http://partsregistry.org/Part:BBa_K398029 BBa_K398029] were equivalent to 33.98% of the Pseudomonas putida aldehyde dehydrogenase activity. Read more about it in our results page
View [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398331 BBa_K398029 in the parts registry]
Specified Components <partinfo>BBa_K398029 SpecifiedComponents</partinfo>
Designer: <partinfo>BBa_K398029 Designer</partinfo>
Status: <partinfo>BBa_K398029 Status</partinfo>
[http://partsregistry.org/Part:BBa_K398331 BBa_K398331] - pCaif
According to our results, the smallest part that we designed it's working!!!
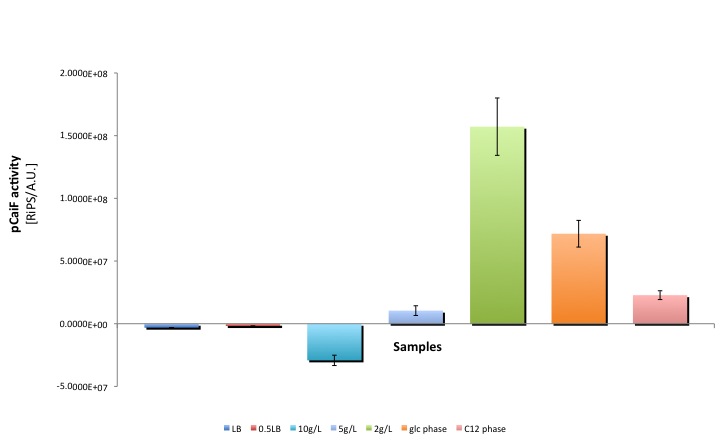
pCaiF a natural promoter in E. coli that is active at high cAMP levels was characterized attaching E00240 in front of our part. Different GFP production rates were found at the stationary phase showing that GFP production depends on the amount of glucose in the medium. Moreover, growth on a secondary carbon source decreases the GFP production rate.
Read more about our fascinating results on our sensing project page
View [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398331 BBa_K398331 in the parts registry]
Specified Components <partinfo>K398331 SpecifiedComponents</partinfo>
Designer: <partinfo>K398331 Designer</partinfo>
Status: <partinfo>K398331 Status</partinfo>
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K398406 BBa_K398406] - Solvent Tolerance
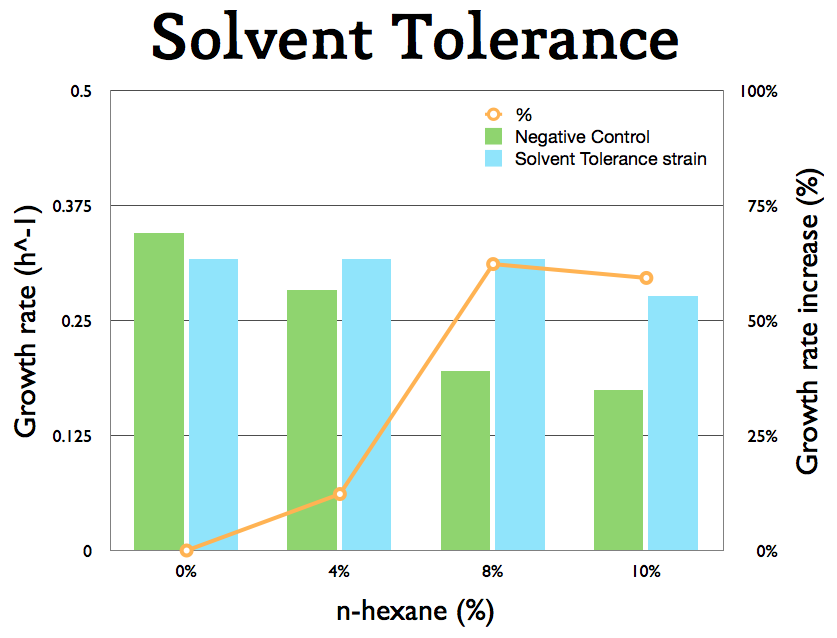
The solvent tolerance cluster (BBa_K398406) was expressed on in E. coli K12. The growth of the cells was tested under different amounts of n-hexane. The results show that this part gives to the cells an growth advantage at higher n-hexane concentrations. The parental strain E. coli K12 grows really slow at 10% (v/v) of n-hexane/M9 mixture.
Read more about these results on our survival project page.
View [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398406 BBa_K398406 in the parts registry]
Specified Components <partinfo>K398406 SpecifiedComponents</partinfo>
Designer: <partinfo>K398406 Designer</partinfo>
Status: <partinfo>K398406 Status</partinfo>
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K398108 BBa_K398108] - Salt Tolerance
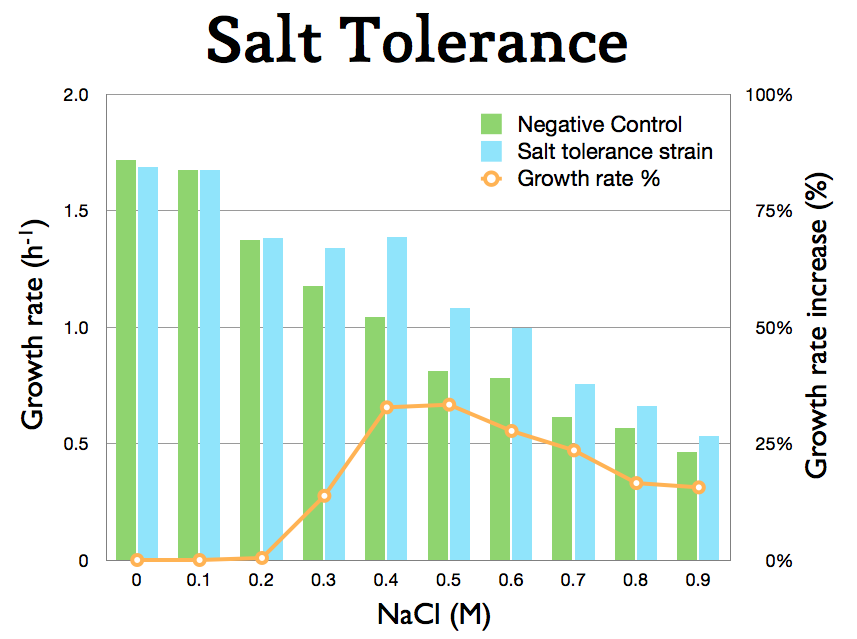
The growth of the bbc1 construct (BBa_K398108) was tested as stated in on the salt tolerance characterization page.
Read more about these results on our survival project page.
View [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398108 BBa_K398108 in the parts registry]
Specified Components <partinfo>K398108 SpecifiedComponents</partinfo>
Designer: <partinfo>K398108 Designer</partinfo>
Status: <partinfo>K398108 Status</partinfo>
BBa_K398206 - Emulsifier
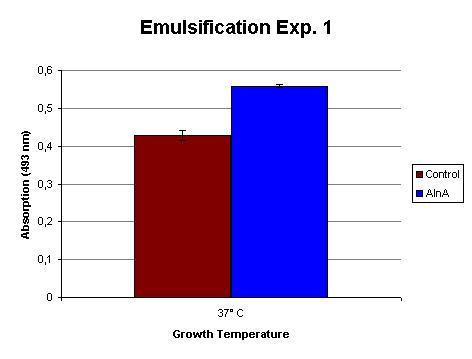
View [http://partsregistry.org/wiki/index.php?title=Part:BBa_K398206 BBa_K398206 in the parts registry]
Specified Components <partinfo>K398206 SpecifiedComponents</partinfo>
Designer: <partinfo>K398206 Designer</partinfo>
Status: <partinfo>K398206 Status</partinfo>
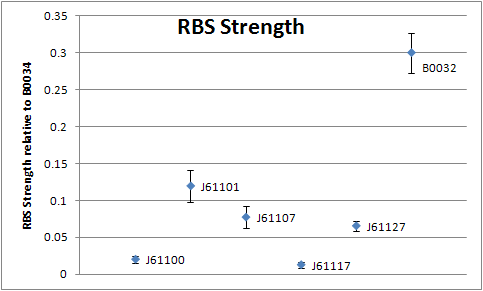
Ribosome Binding Site (RBS) sequence characterization
Read more about the RBS characterization. We used the following 5 constructs to perform the characterization. <partinfo>BBa_K398500 SpecifiedComponents</partinfo> <partinfo>BBa_K398501 SpecifiedComponents</partinfo> <partinfo>BBa_K398502 SpecifiedComponents</partinfo> <partinfo>BBa_K398503 SpecifiedComponents</partinfo> <partinfo>BBa_K398504 SpecifiedComponents</partinfo>
Resulting RBS strenghts:
| Ribosome binding site | Mean relative strength | Standard deviation |
| [http://partsregistry.org/Part:BBa_J61100 J61100] | 0.020 (2.0%) | 0.00512 |
| [http://partsregistry.org/Part:BBa_J61101 J61101] | 0.119 (11.9%) | 0.02140 |
| [http://partsregistry.org/Part:BBa_J61107 J61107] | 0.077 (7.7%) | 0.01480 |
| [http://partsregistry.org/Part:BBa_J61117 J61117] | 0.013 (1.3%) | 0.00448 |
| [http://partsregistry.org/Part:BBa_J61127 J61127] | 0.065 (6.5%) | 0.00660 |
| [http://partsregistry.org/Part:BBa_B0032 B0032] | 0.300 (Reference, 30%) | 0.02690 |
 "
"