Team:Heidelberg/Project/miMeasure
From 2010.igem.org
Laura Nadine (Talk | contribs) |
(→Analysis of Randomized Binding Sites Against Synthetic miRNA) |
||
| Line 27: | Line 27: | ||
===Analysis of Randomized Binding Sites Against Synthetic miRNA=== | ===Analysis of Randomized Binding Sites Against Synthetic miRNA=== | ||
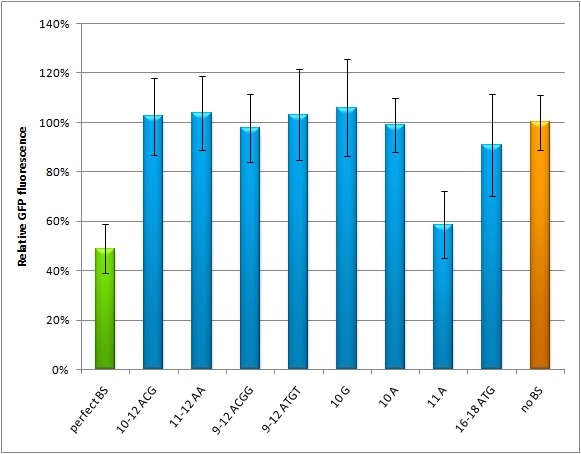
| - | We used microscopy analysis to determine the EGFP expression in relation to EBFP2. EBFP2 serves as a normalization for transfection efficiency. Nine miMeasure constructs with different binding sites were designed. The binding sites are either mutated at one site, or they contain randomly changed sites within a certain range. | + | We used microscopy analysis to determine the EGFP expression in relation to EBFP2. EBFP2 serves as a normalization for transfection efficiency. Nine miMeasure constructs with different binding sites were designed. The binding sites are either mutated at one site, or they contain randomly changed sites within a certain range. If the GFP is down-knocked down completely, it would be 100% of knock-down. The negative control represents 0% knock-down, since there is no binding site cloned into this miMeasure construct. So all the modified binding sites lie in between. The construct, which has the highest efficiency of knock-down carries a perfect binding site, which is complementary to the synthetic miRNA miRsAg. The GFP/BFP-ratio stand for the level of GFP-expression normalized to on copy per cell. We modified binding sites for the synthetic miRNA by introducing random basepairs surrounding the seed region as described above, thereby changing the knockdown efficiency. In figure 1 we plotted the knockdown percentage of our constructs. The miMeasure construct and negative control were [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Transfection co-transfected] with either shRNA miRsAg expressed from a CMV promoter on pcDNA5 or an inert synthetic RNA as control in 1:4 ratio, respectively. |
[[Image:M12-M22_HeLa_daten.jpg|thumb|500px|center|'''GFP/BFP ratio normalized by the negative control''' The data are generated by confocal microscopy of Hela cells, which were transfected with different miMeasure constructs M12-M22 including the negative control (miMeasure construct without binding site). The negative control equals to 1.]] | [[Image:M12-M22_HeLa_daten.jpg|thumb|500px|center|'''GFP/BFP ratio normalized by the negative control''' The data are generated by confocal microscopy of Hela cells, which were transfected with different miMeasure constructs M12-M22 including the negative control (miMeasure construct without binding site). The negative control equals to 1.]] | ||
| Line 81: | Line 81: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
===Analysis of miRaPCR Generated Binding Sites Against a Natural miRNA=== | ===Analysis of miRaPCR Generated Binding Sites Against a Natural miRNA=== | ||
Revision as of 09:43, 27 October 2010

|
|
||||||||||||||||||||||||||||||||||||||||||
 "
"