Team:Heidelberg/Project/miMeasure
From 2010.igem.org
AlejandroHD (Talk | contribs) |
Laura Nadine (Talk | contribs) |
||
| Line 32: | Line 32: | ||
{| class="wikitable sortable" border="0" align="center" style="text-align: left" | {| class="wikitable sortable" border="0" align="center" style="text-align: left" | ||
| - | |-bgcolor=# | + | |-bgcolor=#009be1 |
|+ align="top, left"|'''Table 1: Used Binding Sites and Their Features''' | |+ align="top, left"|'''Table 1: Used Binding Sites and Their Features''' | ||
|sequence||binding site feature||Name/number | |sequence||binding site feature||Name/number | ||
| Line 88: | Line 88: | ||
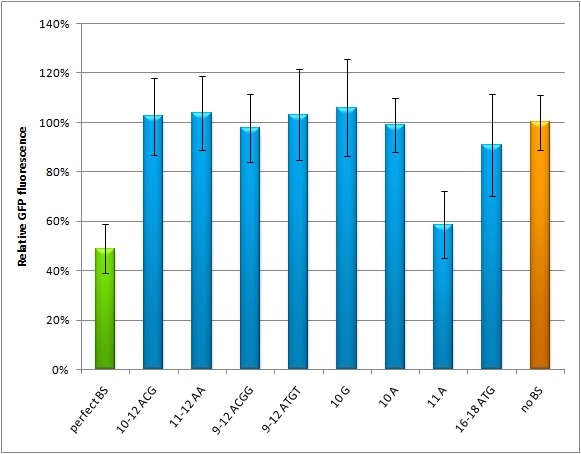
<!--Discussion This experiment shows, that the binding site pattern with 3 aligned perfect binding sites miM-1.3-7 gives the strongest knock-down. Whereby the binding site pattern with two imperfect binding sites 1-8 is weaker, but still stronger than the negative control. If one binding site is randomized from nucleotide 9-12 or 9-22 miM-r12, miM-r22, it loses its capability of protein down-regulation.--> | <!--Discussion This experiment shows, that the binding site pattern with 3 aligned perfect binding sites miM-1.3-7 gives the strongest knock-down. Whereby the binding site pattern with two imperfect binding sites 1-8 is weaker, but still stronger than the negative control. If one binding site is randomized from nucleotide 9-12 or 9-22 miM-r12, miM-r22, it loses its capability of protein down-regulation.--> | ||
{| class="wikitable sortable" border="0" align="center" style="text-align: left" | {| class="wikitable sortable" border="0" align="center" style="text-align: left" | ||
| - | |-bgcolor=# | + | |-bgcolor=#009be1 |
|+ align="top, left"|'''Table2: miRaPCR Designed Binding Sites and Their Features | |+ align="top, left"|'''Table2: miRaPCR Designed Binding Sites and Their Features | ||
|binding site feature'''||Name/number | |binding site feature'''||Name/number | ||
Revision as of 09:27, 27 October 2010

|
|
||||||||||||||||||||||||||||||||||||||||||
 "
"