Team:Heidelberg/Project/miMeasure
From 2010.igem.org
(→Analysis of Randomized Binding Sites Against Synthetic miRNA) |
Laura Nadine (Talk | contribs) |
||
| Line 29: | Line 29: | ||
{| class="wikitable sortable" border="0" align="center" style="text-align: left" | {| class="wikitable sortable" border="0" align="center" style="text-align: left" | ||
|-bgcolor=#cccccc | |-bgcolor=#cccccc | ||
| - | |+ align="top, left"|'''Used Binding Sites and Their Features''' | + | |+ align="top, left"|'''Table 1: Used Binding Sites and Their Features''' |
|sequence||binding site feature||Name/number | |sequence||binding site feature||Name/number | ||
|- | |- | ||
| Line 73: | Line 73: | ||
A control consisting of the empty miMeasure plasmid (without binding site) was also cotransfected with the same conditions a, b, c and d. The cells were used for measurements on day three.--> | A control consisting of the empty miMeasure plasmid (without binding site) was also cotransfected with the same conditions a, b, c and d. The cells were used for measurements on day three.--> | ||
| - | ===Analysis of | + | ===Analysis of miRaPCR Generated Binding Sites Against a Natural miRNA=== |
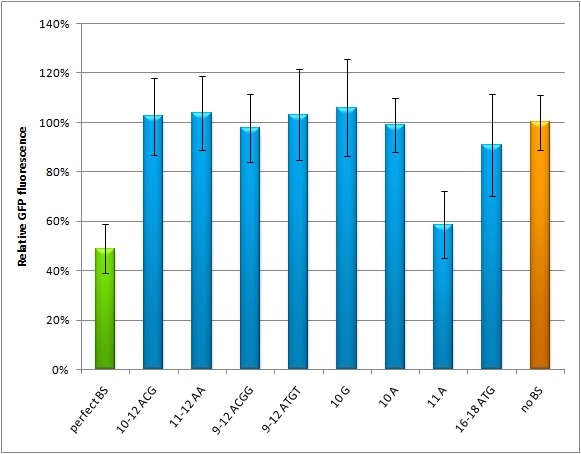
| - | The | + | The [https://2010.igem.org/Team:Heidelberg/Notebook/BSDesign/July miRaPCR] generates binding sites out of rationally designed fragments. These are aligned with each other by chance, whereby different spacer regions are inserted randomly in between. It has been suggested that having more than one binding site of for the same miRNA behind each other can lead to stronger down-regulation than a single one. If imperfect binding sites are aligned, it is also supposed to be stronger than a single one. This is what we tested using MiRaPCR for effortless assembly of binding site fragments. |
| - | + | For our experiments, we took advantage of the high abundance of miRNA 122 in liver cells and tested different combinations of binding sites created by miRaPCR. We transfected HeLa and HuH7 cells with the constructs described in table 2. Since HuH7 cells express miR-122, the constructs will be affected in the HuH7 cells without cotransfecting any miRNAs, whereas miR-122 were cotransfected for the HeLa cells in 2:1 ratio. The result shows the ratio between GFP and BFP normalized to the negative control (miMeasure constructs without binding sites). The miMeasure constructs where also compared to the expression of miMeasure containing one perfect binding site for miRNA 122. | |
| - | + | <!—Discussion This experiment shows, that the binding site pattern with 3 aligned perfect binding sites miM-1.3-7 gives the strongest knock-down. Whereby the binding site pattern with two imperfect binding sites 1-8 is weaker, but still stronger than the negative control. If one binding site is randomized from nucleotide 9-12 or 9-22 miM-r12, miM-r22, it loses its capability of protein down-regulation.--> | |
| - | + | {| class="wikitable sortable" border="0" align="center" style="text-align: left" | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| class="wikitable sortable" border="0" style="text-align: left" | + | |
|-bgcolor=#cccccc | |-bgcolor=#cccccc | ||
| - | |+ align="top, left"|''' | + | |+ align="top, left"|'''Table2: miRaPCR Designed Binding Sites and Their Features |
|binding site feature'''||Name/number | |binding site feature'''||Name/number | ||
|- | |- | ||
Revision as of 09:13, 27 October 2010

|
|
||||||||||||||||||||||||||||||||||||||||||
 "
"