Team:Heidelberg/Project/miMeasure
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Laura Nadine (Talk | contribs) |
||
| Line 23: | Line 23: | ||
===Microscopy=== | ===Microscopy=== | ||
| - | + | ===Analysis of mutated/ randomized binding sites against synthetic miRNA=== | |
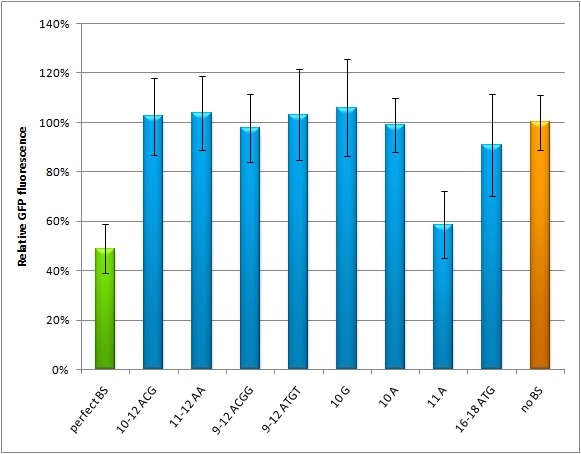
We used microscopy analysis to determine the EGFP expression in relation to EBFP2. EBFP2 serves as a normalization for transfection efficiency. Nine miMeasure constructs with different binding sites were designed. The binding sites are either mutated at one site, or they contain randomly changed sites within a certain range. The construct representing the 100% knock-down is the perfect binding site, which is complementary to the synthetic miRNA miRsAg. The negative control represents 0% knock-down, since there is no binding site cloned into this miMeasure construct. M12 contains the perfect binding site. The GFP/BFP-ratio stand for the level of GFP-expression normalized to one copy per cell. When we compare the GFP/BFP-ratio between the constructs, there is a significant difference of GFP expression in the control (miMeasure without binding site) and the construct containing the perfect binding site for the cotransfected synthetic miRNA. The modified binding sites don't supress GFP-expression as much as the perfect one. So GFP expression is just in between, except for constructs M20 and M22. | We used microscopy analysis to determine the EGFP expression in relation to EBFP2. EBFP2 serves as a normalization for transfection efficiency. Nine miMeasure constructs with different binding sites were designed. The binding sites are either mutated at one site, or they contain randomly changed sites within a certain range. The construct representing the 100% knock-down is the perfect binding site, which is complementary to the synthetic miRNA miRsAg. The negative control represents 0% knock-down, since there is no binding site cloned into this miMeasure construct. M12 contains the perfect binding site. The GFP/BFP-ratio stand for the level of GFP-expression normalized to one copy per cell. When we compare the GFP/BFP-ratio between the constructs, there is a significant difference of GFP expression in the control (miMeasure without binding site) and the construct containing the perfect binding site for the cotransfected synthetic miRNA. The modified binding sites don't supress GFP-expression as much as the perfect one. So GFP expression is just in between, except for constructs M20 and M22. | ||
| Line 31: | Line 31: | ||
[[Image:M12-M22_HeLa_daten.jpg|thumb|500px|center|'''GFP/BFP ratio normalized by the negative control''' The data are generated by confocal microscopy of Hela cells, which were transfected with different miMeasure constructs M12-M22 including the negative control (miMeasure construct without binding site). The negative control equals to 1.]] | [[Image:M12-M22_HeLa_daten.jpg|thumb|500px|center|'''GFP/BFP ratio normalized by the negative control''' The data are generated by confocal microscopy of Hela cells, which were transfected with different miMeasure constructs M12-M22 including the negative control (miMeasure construct without binding site). The negative control equals to 1.]] | ||
| - | ===Analysis of binding sites against synthetic miRNA miRsAg=== | + | ====Analysis of binding sites against synthetic miRNA miRsAg==== |
5000 HeLa cells were seeded on day one in each well of the 96 well plate. Transfection of the constructs (M12-M22) with four different conditions were carried out on day two. The ratio of transfection is 1 (M construct) : 5 (stuffer/ miRsAg/ pcDNA5/ shRNA3) with a total amount of 50ng DNA. | 5000 HeLa cells were seeded on day one in each well of the 96 well plate. Transfection of the constructs (M12-M22) with four different conditions were carried out on day two. The ratio of transfection is 1 (M construct) : 5 (stuffer/ miRsAg/ pcDNA5/ shRNA3) with a total amount of 50ng DNA. | ||
| Line 47: | Line 47: | ||
{| class="wikitable sortable" border="0" style="text-align: left" | {| class="wikitable sortable" border="0" style="text-align: left" | ||
|-bgcolor=#cccccc | |-bgcolor=#cccccc | ||
| - | |+ align="top, left"|''' | + | |+ align="top, left"|'''Used Binding Sites and Their Features''' |
|sequence||binding site feature||Name/number | |sequence||binding site feature||Name/number | ||
|- | |- | ||
| Line 85: | Line 85: | ||
{| class="wikitable sortable" border="0" style="text-align: left" | {| class="wikitable sortable" border="0" style="text-align: left" | ||
|-bgcolor=#cccccc | |-bgcolor=#cccccc | ||
| - | |+ align="top, left"|''' | + | |+ align="top, left"|'''MiRaPCR Designed Binding Sites and Their Features |
|binding site feature'''||Name/number | |binding site feature'''||Name/number | ||
|- | |- | ||
Revision as of 07:39, 27 October 2010

|
|
||||||||||||||||||||||||||||||||||||||||||
 "
"