Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→Introduction) |
(→Introduction) |
||
| Line 24: | Line 24: | ||
==Introduction== | ==Introduction== | ||
| - | MicroRNAs (miRNAs) are short endogenous, non-coding RNAs that mediate gene expression in a diversity of organisms {{HDref|Bartel, 2004}}. Although the understanding of their biological functions is progressing remarkably, the exact mechanisms of regulation are still not unambiguously defined. However, it is commonly believed that miRNAs '''trigger target mRNA regulation''' by binding to 3’ untranslated region (UTR) of its target {{HDref|Chekulaeva and Filipowicz, 2009}}. <!--The discovery of the first miRNA (lin-4) revealed sequence complementarity to multiple conserved sites in the 3’UTR of the lin-14 mRNA {{HDref|Lee et al., 1993; Wightman et al., 1993}}. --> Exact principles of expression knockdown mediated by miRNA are still in debate {{HDref|Eulalio et al., 2008}}.<br/>However, sequence depending '''binding site properties''' have an essential impact on miRNA-mRNA interaction. <!--[figure, short explanations on seed regions, flanking regions, spacers, mismatches and resulting bulges]. Some functionally important sections of miRNAs have been described in literature, such as the seed region {{HDref|Grimson et al., 2007; Bartel, 2009}}. It is defined as a miRNA region of seven nucleotides length that shows perfect pairing the mRNA target sequence. --><!--The seed usually consists of the nucleotides on position 2-8 of a miRNA binding sites in the 5'UTR of the mRNA. Based on this simple principle, we randomized our miRNA binding sites between nucleotide 9 - 12 or 9 - 22 in the so called flanking region. Alternatively, we tried rational exchanges of nucleotides to see how they effect binding of the miRNA to its target mRNA. --> Depending on pairing specificity translational repression is mediated through the imperfect miRNA-mRNA hybrids. The potential for stringent regulation of transgene expression makes the miRNA world a promising area of gene therapy {{HDref|Brown et al.,2009}}. There is a need for tight control of gene expression, since cellular processes are sensitive to expression profiles. Non-mediated gene expression can lead to fatal dysfunction of molecular networks. It is widely known, that miRNAs can adjust such fluctuations{{HDref|Brenecke et al., 2005}}. A combination of random and rational '''design''' of binding sites could become a '''powerful tool''' to achieve a narrow range of resulting gene expression knockdown. To ease <i>in silico</i> construction of miRNA binding sites with appropriate characteristics for its target, we wrote a program - the [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer]. Using all of our [https://2010.igem.org/Team:Heidelberg/Modeling theoretical models] gives the user the opportunity to calculate knockdown percentages caused by the designed miRNA in the target cell.<!--The experimental applicability is still limited by redundant target sites and various miRNA expression patterns within the cells. This hampers distinct expression levels of the gene of interest (GOI) fused to the miRNA binding site.--> | + | MicroRNAs (miRNAs) are short endogenous, non-coding RNAs that mediate gene expression in a diversity of organisms {{HDref|Bartel, 2004}}. Although the understanding of their biological functions is progressing remarkably, the exact mechanisms of regulation are still not unambiguously defined. However, it is commonly believed that miRNAs '''trigger target mRNA regulation''' by binding to 3’ untranslated region (UTR) of its target {{HDref|Chekulaeva and Filipowicz, 2009}}. <!--The discovery of the first miRNA (lin-4) revealed sequence complementarity to multiple conserved sites in the 3’UTR of the lin-14 mRNA {{HDref|Lee et al., 1993; Wightman et al., 1993}}. --> Exact principles of expression knockdown mediated by miRNA are still in debate {{HDref|Eulalio et al., 2008}}.<br/>However, sequence depending '''binding site properties''' have an essential impact on miRNA-mRNA interaction. <!--[figure, short explanations on seed regions, flanking regions, spacers, mismatches and resulting bulges]. Some functionally important sections of miRNAs have been described in literature, such as the seed region {{HDref|Grimson et al., 2007; Bartel, 2009}}. It is defined as a miRNA region of seven nucleotides length that shows perfect pairing the mRNA target sequence. --><!--The seed usually consists of the nucleotides on position 2-8 of a miRNA binding sites in the 5'UTR of the mRNA. Based on this simple principle, we randomized our miRNA binding sites between nucleotide 9 - 12 or 9 - 22 in the so called flanking region. Alternatively, we tried rational exchanges of nucleotides to see how they effect binding of the miRNA to its target mRNA. --> Depending on pairing specificity translational repression is mediated through the imperfect miRNA-mRNA hybrids. The potential for stringent regulation of transgene expression makes the miRNA world a promising area of gene therapy {{HDref|Brown et al.,2009}}. There is a need for tight control of gene expression, since cellular processes are sensitive to expression profiles. Non-mediated gene expression can lead to fatal dysfunction of molecular networks. It is widely known, that miRNAs can adjust such fluctuations {{HDref|Brenecke et al., 2005}}. A combination of random and rational '''design''' of binding sites could become a '''powerful tool''' to achieve a narrow range of resulting gene expression knockdown. To ease <i>in silico</i> construction of miRNA binding sites with appropriate characteristics for its target, we wrote a program - the [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer]. Using all of our [https://2010.igem.org/Team:Heidelberg/Modeling theoretical models] gives the user the opportunity to calculate knockdown percentages caused by the designed miRNA in the target cell.<!--The experimental applicability is still limited by redundant target sites and various miRNA expression patterns within the cells. This hampers distinct expression levels of the gene of interest (GOI) fused to the miRNA binding site.--> |
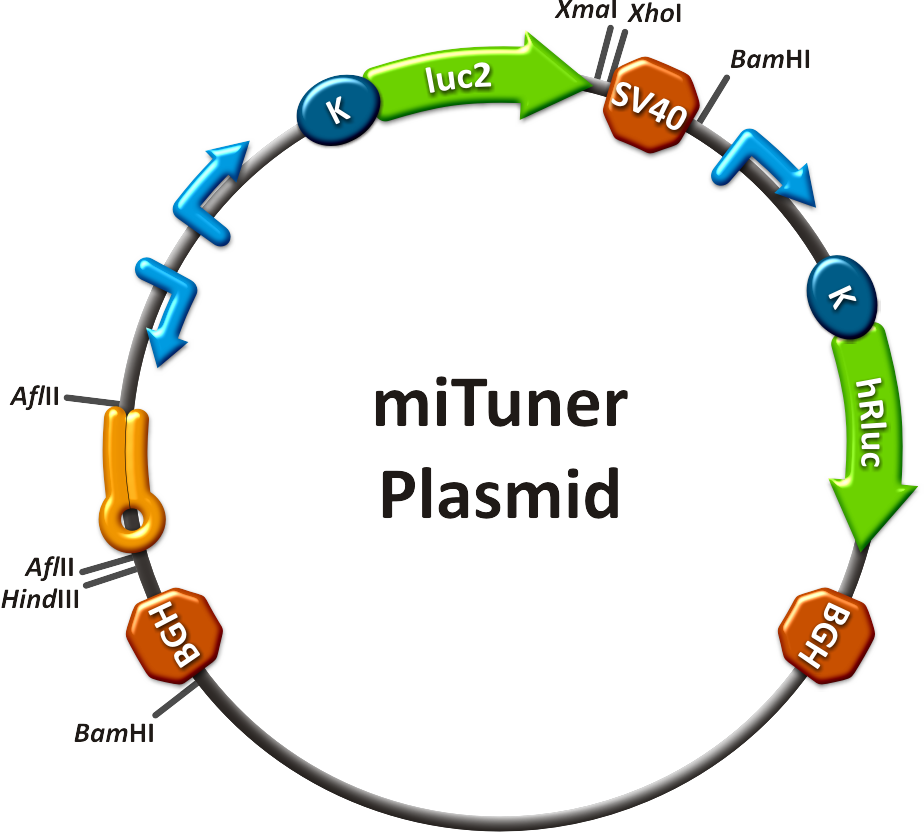
Our '''synthetic miRNA Kit''' guarantees at least for individually modifiable but still ready-to-use constructs to interfere genetic circuits with synthetic or endogenous miRNAs. We preciously show, that gene expression can thereby by adjusted - tuned - to an arbitrary level. The '''miTuner''' (see sidebar) allows on the simultaneous expression of a synthetic miRNA and a gene of interest that is fused with a designed binding site for this specific miRNA. Our modular kit comes with different parts that can be combined by choice, e. g. different mammalian promoters and characterized binding sites of specific properties. By choosing a certain binding site to tag the GOI, one can tune the expression of this gene. Depending on the GOI, different means for read out of gene expression come into play. At first, we applied [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay dual-luciferase assay], since we used Luciferase as a reporter for a proof-of-principle approach. Later on, semi-quantitative immunoblots were prepared for testing of therapeutic genes. However, all the received information fed our models, thereby creating an '''integrative feedback loop between experiments and simulation'''. | Our '''synthetic miRNA Kit''' guarantees at least for individually modifiable but still ready-to-use constructs to interfere genetic circuits with synthetic or endogenous miRNAs. We preciously show, that gene expression can thereby by adjusted - tuned - to an arbitrary level. The '''miTuner''' (see sidebar) allows on the simultaneous expression of a synthetic miRNA and a gene of interest that is fused with a designed binding site for this specific miRNA. Our modular kit comes with different parts that can be combined by choice, e. g. different mammalian promoters and characterized binding sites of specific properties. By choosing a certain binding site to tag the GOI, one can tune the expression of this gene. Depending on the GOI, different means for read out of gene expression come into play. At first, we applied [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay dual-luciferase assay], since we used Luciferase as a reporter for a proof-of-principle approach. Later on, semi-quantitative immunoblots were prepared for testing of therapeutic genes. However, all the received information fed our models, thereby creating an '''integrative feedback loop between experiments and simulation'''. | ||
Revision as of 01:20, 27 October 2010

|
|
||
 "
"