Team:Alberta/biobyte2
From 2010.igem.org
Rpgguardian (Talk | contribs) |
Rpgguardian (Talk | contribs) |
||
| Line 18: | Line 18: | ||
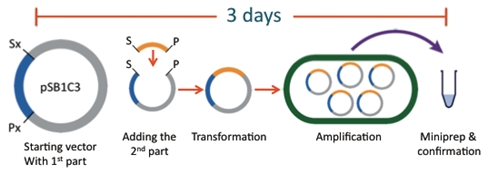
| - | [[Image:team-alberta-biobrick-tour.jpg|center|frame|Traditional BioBrick construction.]] | + | [[Image:team-alberta-biobrick-tour.jpg|center|frame|Figure 1.1. Traditional BioBrick construction which takes approximately 3 days to complete from the cutting of the original constructs to transformation and confirmation.]] |
| + | |||
====Comparison==== | ====Comparison==== | ||
| + | |||
| + | |||
==BioBytes Vs BioBytes 2.0== | ==BioBytes Vs BioBytes 2.0== | ||
<div id="horiz-line"></div> | <div id="horiz-line"></div> | ||
Revision as of 23:03, 26 October 2010
Overview
BioBytes 2.0 is the heart of the GENOMIKON kit. We have taken the original BioBytes Assembly System developed from last year's iGEM project and adapted it for a high school setting. The system is designed by the sequential addition of gene elements with alternating ends bla ble bla bla bla...
Traditional Methods
The current assembly standard is the BioBrick method. While the registry of parts and the assembly standard has allowed for efficient construction of plasmids in a laboratory setting, it has numerous limitations prohibiting its use in high schools. For example, common laboratory protocols such as transformation, ligation, and restriction digestion require materials and equipment not available to high schools. Not only does this require expensive reagents and equipment, it also takes days to weeks to assemble acomplicated construct. An experiment of such length far surpasses the average high school student’s attention span and the time a curriculum can spend on a particular subject.
Comparison
BioBytes Vs BioBytes 2.0
Components of the System
Anchor
BioBytes
Cap
Byte Construction
Assembly System
The traditional "BioBrick" construction method, while well suited to simple plasmid construction tasks, is a very time consuming process when larger, more complicated plasmids are attempted. Each cycle of adding a part to a BioBrick backbone takes at least three days.
The "BioBytes Version 2.0" construction method has been shown to create (insert actual data here) plasmids from up to 8 separate parts in
an afternoon's work. This is a vast improvement.
[[Image:team-alberta-building-tour.jpg|center|frame|BioByte version 2.0 construction.]
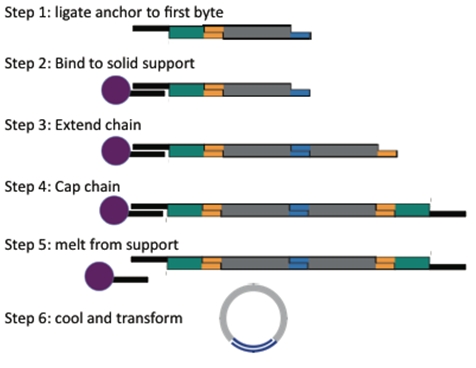
BioBytes Components
The method has three main components:
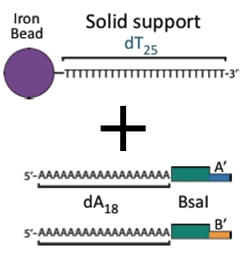
The Anchor
A ferro-magnetic bead attached to a piece of DNA. This piece serves as the initial piece from which we assemble a DNA construct. The bead allows us to manipulate the DNA with magnets making washing and subsequent attachments easier.
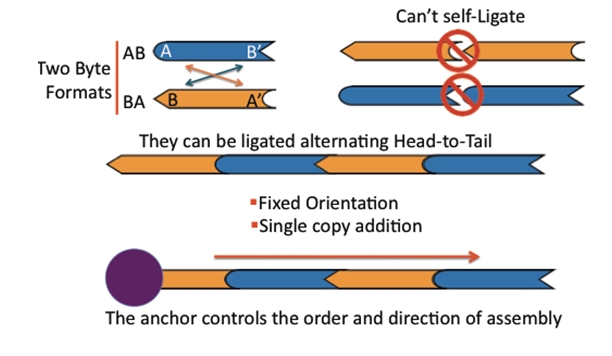
The BioBytes
DNA fragments that can be attached together to build up a larger construct. There are two types of pieces, AB and BA. The A end can join only with another A end and the B end can join only with another B end. As a result pieces can only be joined in a single orientation.
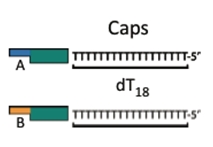
The Cap
A DNA fragment that finishes off a construct and allows for circularization of the construct into a plasmid.
The process of building a plasmid is more elegant and more rapid than the current biobyte system!
The Process
Starting with an anchor, add the first piece and ligate.
Then hold the single piece construct in the tube by placing it on the magnetic rack. Now you can wash away most of the excess of piece 1.
Add the next piece and repeat until you have added all the pieces you want.
Then add the cap.
Now just heat to release the anchor and open up the cap, upon cooling the construct will circularize.
Easy!
Using this process we were able to assemble eight pieces in an afternoon!
 "
"