Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→Introduction) |
(→planing) |
||
| Line 28: | Line 28: | ||
<br/> | <br/> | ||
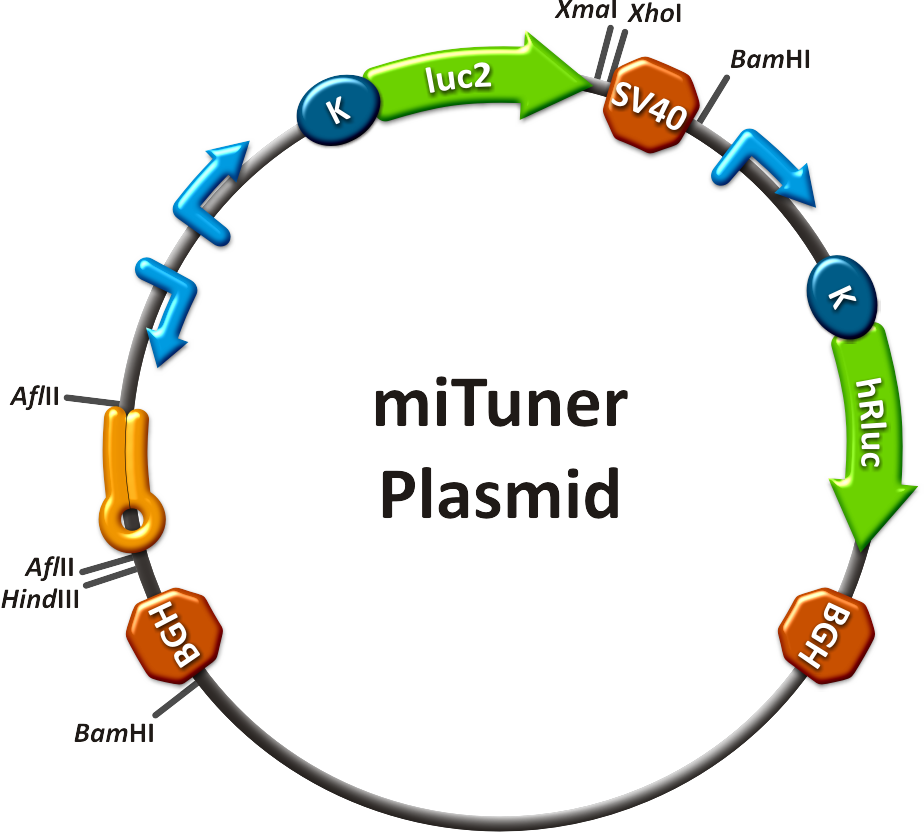
=== planing === | === planing === | ||
| - | A combination of random and rational '''design''' of binding sites could become a '''powerful tool''' to achieve a narrow range of resulting gene expression knockdown. To ease <i>in silico</i> construction of miRNA binding sites with appropriate characteristics for its target, we wrote a program - the [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer]. | + | A combination of random and rational '''design''' of binding sites could become a '''powerful tool''' to achieve a narrow range of resulting gene expression knockdown. To ease <i>in silico</i> construction of miRNA binding sites with appropriate characteristics for its target, we wrote a program - the [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer]. Using all of our [https://2010.igem.org/Team:Heidelberg/Modeling theoretical models] gives the user the opportunity to calculate knockdown percentages caused by the designed miRNA in the target cell. |
=== experiments === | === experiments === | ||
Revision as of 21:58, 26 October 2010

|
|
||
 "
"