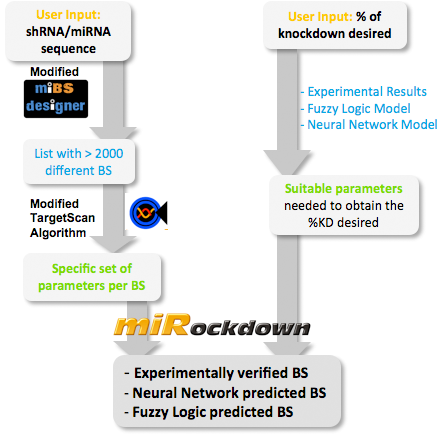
Team:Heidelberg/Modeling/descriptions
From 2010.igem.org
(→miRockdown) |
(→miBS designer) |
||
| Line 23: | Line 23: | ||
</html> | </html> | ||
| - | ==miBS designer== | + | ==miBSdesigner== |
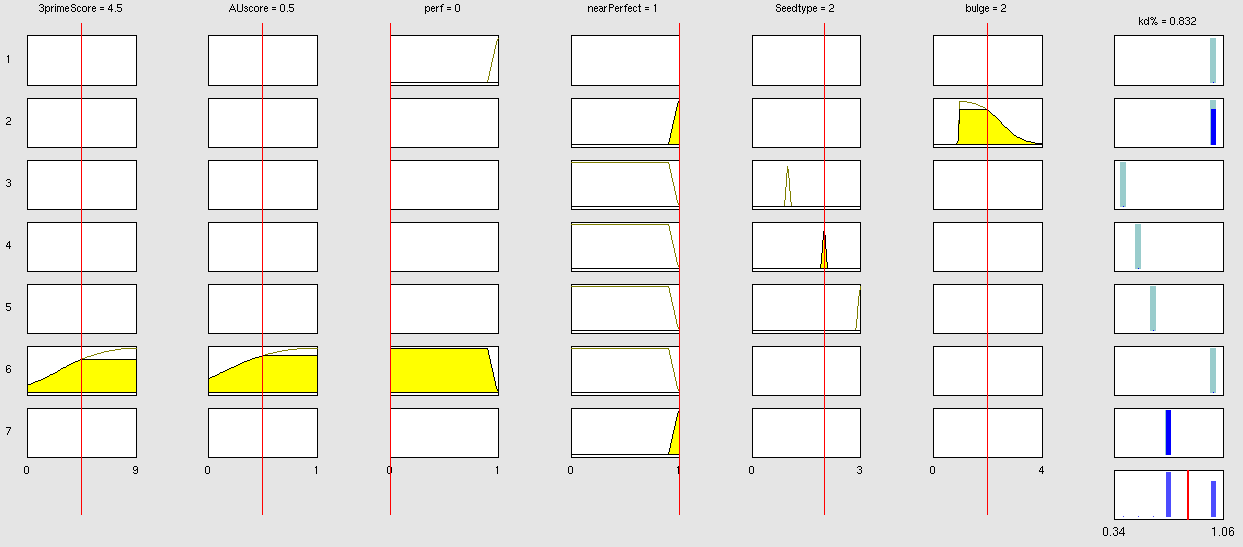
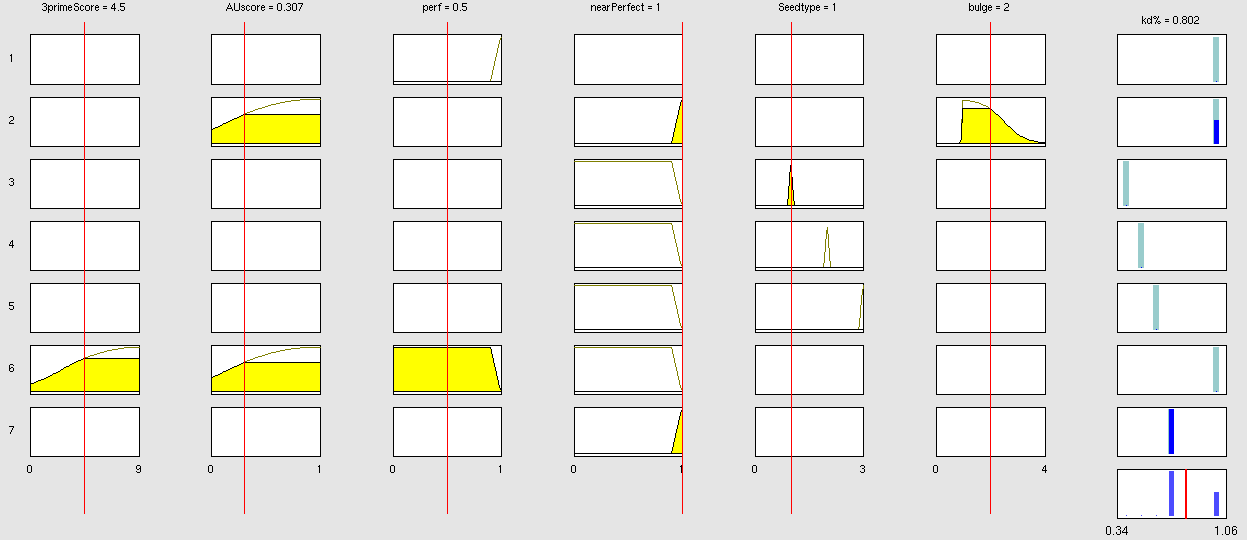
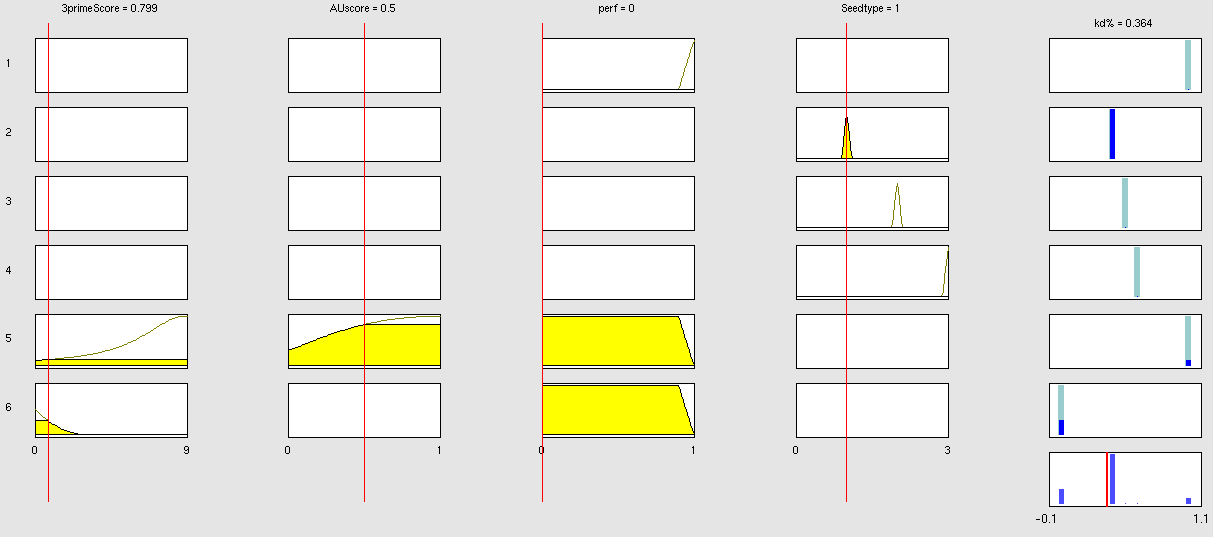
| + | Very early we realized that having a binding site designer was crucial to complete the computational approach to our project: miBSdesigner is an easy-to-use application to create in silico binding sites for any given miRNA. By using our device, the user will be able to generate binding sites with several different properties. | ||
| + | |||
| + | ===Input=== | ||
| + | The user has to input a name for the miRNA to name the primers. The miRNA sequence must be 22 nucleotides long and has to be input in direction 5’ to 3’ (both DNA and RNA sequences are admitted and any extra characters will be removed from the sequence). The user can also enter a spacer inert sequence if he needs to place the binding site further along in the 3’UTR region (it is recommended that the binding site is at least 15 nucleotides away from the stop codon).Initially the user can choose between a perfect binding site (matching the 22 nucleotides), or an almost perfect binding site (matching all of the nucleotides, but leaving a 4-nucleotide bulge between 9 and 12. | ||
| + | Apart from these two options, the user can personalize the binding site to meet their individual requirements. | ||
| + | |||
| + | ===Seed types=== | ||
| + | [[Image:MiRNA_BS_examples_small.jpg]] | ||
| + | |||
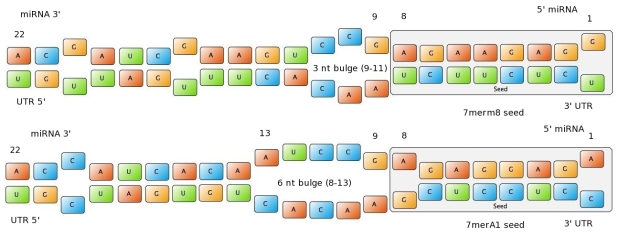
| + | Figure 1: Interactions between two miRNAs and their binding sites. Examples to show different types of seeds. | ||
| + | |||
| + | |||
| + | In miBS designer, the user can choose between several types of seed for their binding site (ordered by increasing efficacy): | ||
| + | |||
| + | - 6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. | ||
| + | |||
| + | - 7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. | ||
| + | |||
| + | - 7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. | ||
| + | |||
| + | - 8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. | ||
| + | |||
| + | - Apart from any of these options, the user can decide to create a customized seed with a mismatch included. | ||
| + | |||
| + | The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | ||
| + | |||
| + | ===Supplementary region=== | ||
| + | In miBS designer, the user can choose among several types of supplementary sequences, starting with 3 matching nucleotides (14-16), increasing sequentially until 8 (13-20), and then total matching (from 13-22, leaving a bulge). In case the user needs some other specific supplementary region, he can customize the sequence by inputting the desired matching nucleotides. | ||
| + | |||
| + | ===AU content=== | ||
| + | In order to allow the user to improve the efficacy of their binding sites, miBS designer offers options to increase the AU content by adding adenine or uracil to positions around the matches (specifically in -1, 0, 1, 8, 9 and 10). The function is designed so that it varies the AU content without introducing new pairings. | ||
| + | |||
| + | ===Sticky ends=== | ||
| + | In order to facilitate the task of introducing the binding site into a plasmid, the user can add sequences to both ends of the binding site. Initially, the user can choose among the RFC-12 standard for biobricks BB2, the XmaI/XhoI restriction enzymes used IN WHAT??, or some custom sequences input by the user. In the last case, the output sequences will not be directly ready for cloning: the user has to either digest the construction prior to ligation, or to process the primers before ordering them to remove the extra nucleotides. | ||
| + | |||
| + | ===Output=== | ||
| + | miBS designer generates the primer needed to integrate the binding site desired, into a plasmid, alongside with the primer for the complementary strand. It will also produce specific names for the two primers. | ||
| + | |||
==miCrappallo== | ==miCrappallo== | ||
Revision as of 21:54, 26 October 2010

|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"