Team:Uppsala-SwedenWeek15
From 2010.igem.org
(→Plasmid concentration of K1, K2, K3, (old version) and digestion) |
(→Plasmid concentration of K1, K2, K3, (old version) and digestion) |
||
| Line 866: | Line 866: | ||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
| + | |||
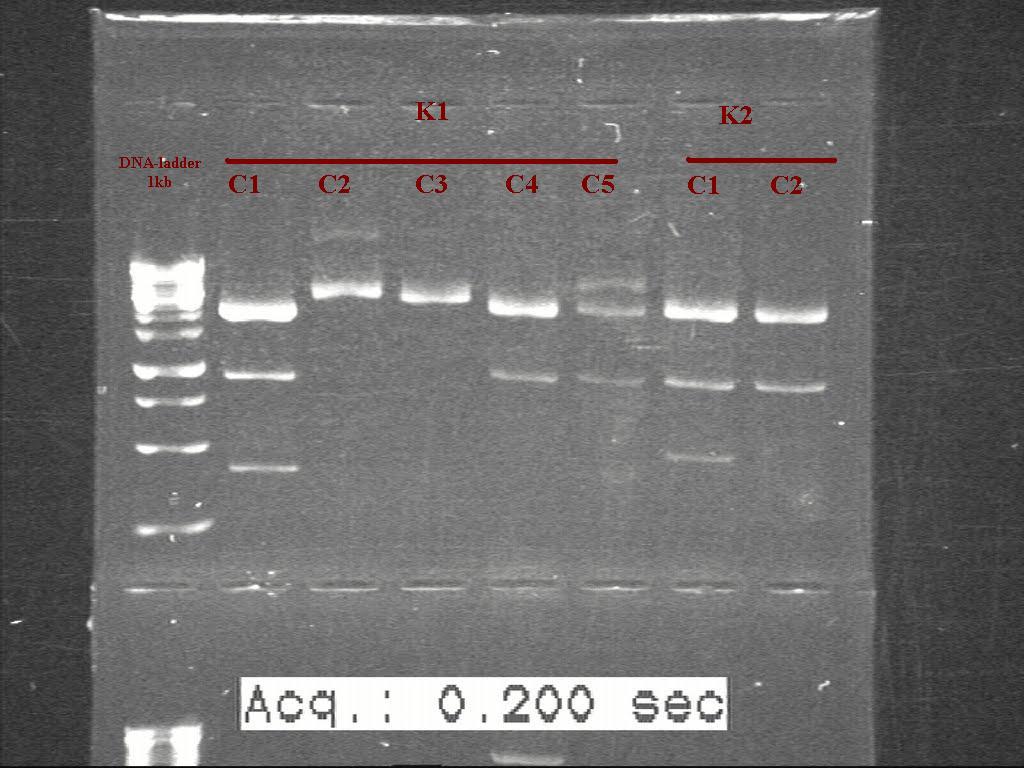
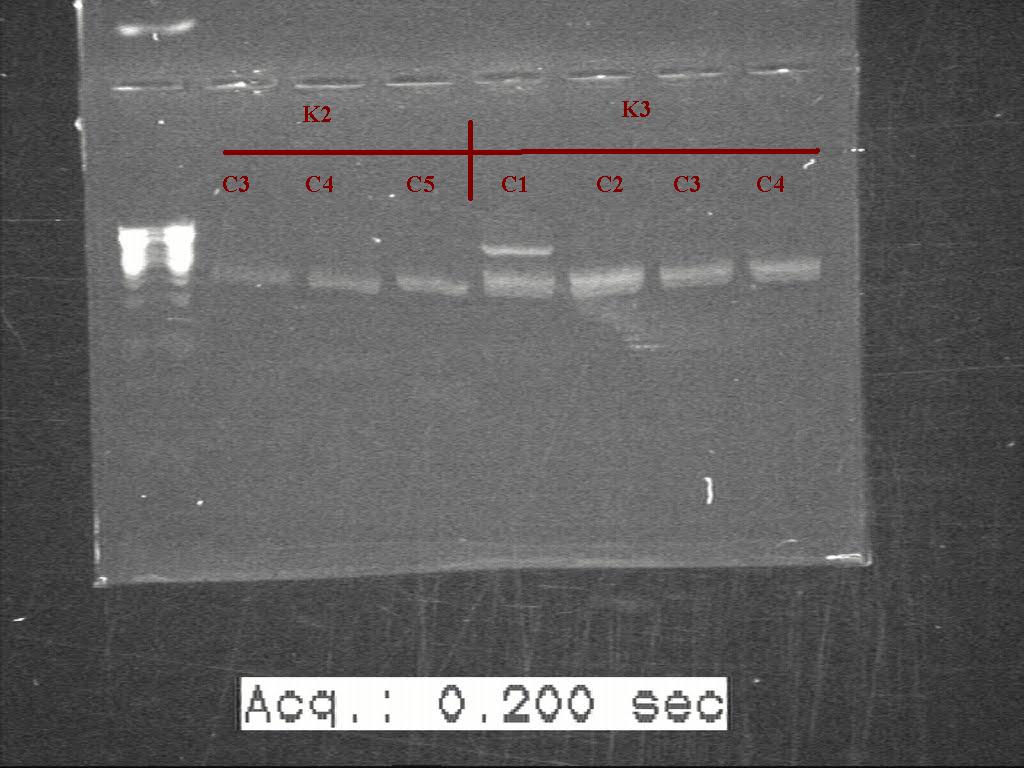
| + | [[Image:Labnotes html m15593582.jpg|500px|thumb|left|K1 K2]] [[Image:Labnotes html m4d07be2b.jpg|500px|thumb|right|K2 K3]] | ||
| + | |||
| + | |||
| + | The K4, K5 and K6 construct are composed of J1, J2 and J3 assembled with BB_I0460 which contains both Ampicillin and Kanamycin cassettes in its backbone, psB1AK3, and the J- construct contains Chloramphinicol cassette, psB1C3. The Tetracyclin antibiotic hasn’t work as expected. Therefore the 3A assembly technique wouldn’t be possible to apply due there is no antibiotics left available with a corresponding backbone. | ||
| + | |||
| + | One option to get around this obstacle was to run the gel, after digestion of BB_I04060, and cut the band with the Biobrick-length and then 3A assembly it with the J-construct and E-P digested psB1C3. | ||
| + | |||
| + | Another option was to switch backbone of J-construct to psB1C3 and then use 3A-assembly it with BB_I0460 and E-P digested psB1C3. We used the second method because it’s a more safer way even though it takes longer time to finish. | ||
Revision as of 19:36, 26 October 2010



Week-15
Plasmid concentration of K1, K2, K3, (old version) and digestion
|
Sample |
DNA-concentration (µL/ml) |
DNA |
Enzymes (E & P) |
FDB (10X) |
H20 |
Total (µL) |
|
K1C1 |
32,7 |
15.3 |
0,5 &0.5 |
2 |
1.7 |
20 |
|
K1C2 |
98.3 |
5.0 |
0,5 &0.5 |
2 |
12 |
20 |
|
K1C3 |
74.4 |
6.7 |
0,5 &0.5 |
2 |
10.3 |
20 |
|
K1C4 |
516.4 |
0.9 |
0,5 &0.5 |
1 |
7.1 |
10 |
|
K1C5 |
135.4 |
3.6 |
0,5 &0.5 |
1 |
4.4 |
10 |
|
K2C1 |
161.7 |
3.1 |
0,5 &0.5 |
1 |
4.9 |
10 |
|
K2C2 |
92.3 |
5.4 |
0,5 &0.5 |
1 |
2.6 |
10 |
|
K2C3 |
112.2 |
4.3 |
0,5 &0.5 |
1 |
3.7 |
10 |
|
K2C4 |
45.6 |
10.9 |
0,5 &0.5 |
2 |
6.1 |
20 |
|
K2C5 |
348.1 |
1.4 |
0,5 &0.5 |
1 |
6.6 |
10 |
|
K2C6 |
91.3 |
5.4 |
0,5 &0.5 |
1 |
2.6 |
10 |
|
K3C1 |
88.4 |
5.6 |
0,5 &0.5 |
1 |
2.4 |
10 |
|
K3C2 |
109.9 |
4.5 |
0,5 &0.5 |
1 |
3.5 |
10 |
|
K3C3 |
116.6 |
4.3 |
0,5 &0.5 |
1 |
3.8 |
10 |
|
K3C4 |
110.0 |
4.6 |
0,5 &0.5 |
1 |
3.5 |
10 |
|
K3C5 |
90.67 |
5.5 |
0,5 &0.5 |
1 |
2.5 |
10 |
|
K3C6 |
109.5 |
4.6 |
0,5 &0.5 |
1 |
3.4 |
10 |
E= EcorI
P= PstI
FDB= Fast Digest Buffer
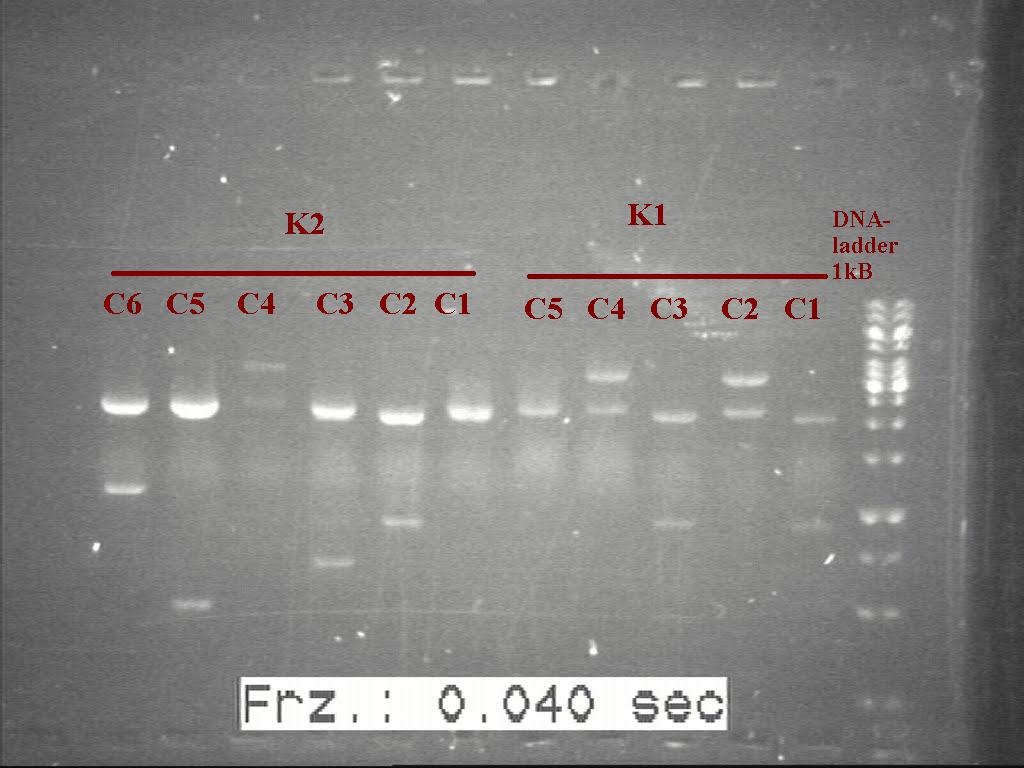
Gel of digested samples with 0.8% agarose gel . picture missing
Counting from the ladder. K1(Colony 1-5) and K2(colony 1-6)
|
Sample |
DNA (µL/ml) |
DNA 100X) |
DNA required |
Enzymes (E & P) |
FDB (10X) |
H20 |
Total (µL) |
|
K1C1 |
0.941 |
94.1 |
5.3 |
0,5 &0.5 |
1 |
2.7 |
20 |
|
K1C2 |
0.670 |
67.0 |
7.4 |
0,5 &0.5 |
1 |
0.6 |
20 |
|
K1C3 |
0.736 |
73.6 |
6.7 |
0,5 &0.5 |
1 |
1.3 |
20 |
|
K1C4 |
0.522 |
52.2 |
9.5 |
0,5 &0.5 |
2 |
7.5 |
10 |
|
K1C5 |
0.622 |
62.2 |
8.0 |
0,5 &0.5 |
2 |
9.0 |
10 |
|
K2C1 |
0.806 |
80.6 |
6.2 |
0,5 &0.5 |
1 |
1.8 |
10 |
|
K2C2 |
0.567 |
56.7 |
8.8 |
0,5 &0.5 |
2 |
2.6 |
10 |
|
K2C3 |
0.611 |
61.1 |
8.1 |
0,5 &0.5 |
2 |
3.7 |
10 |
|
K2C4 |
0.671 |
67.1 |
7.4 |
0,5 &0.5 |
1 |
6.1 |
20 |
|
K2C5 |
0.828 |
82.8 |
6.0 |
0,5 &0.5 |
1 |
6.6 |
10 |
|
K3C1 |
0.678 |
67.8 |
7.3 |
0,5 &0.5 |
1 |
2.6 |
10 |
|
K3C2 |
0.955 |
95.5 |
5.2 |
0,5 &0.5 |
1 |
2.4 |
10 |
|
K3C3 |
0.841 |
84.1 |
5.9 |
0,5 &0.5 |
1 |
3.5 |
10 |
|
K3C4 |
1.11 |
111.0 |
4.5 |
0,5 &0.5 |
1 |
3.8 |
10 |
|
K3C5 |
0.868 |
86.8 |
5.7 |
0,5 &0.5 |
1 |
3.5 |
10 |
The K4, K5 and K6 construct are composed of J1, J2 and J3 assembled with BB_I0460 which contains both Ampicillin and Kanamycin cassettes in its backbone, psB1AK3, and the J- construct contains Chloramphinicol cassette, psB1C3. The Tetracyclin antibiotic hasn’t work as expected. Therefore the 3A assembly technique wouldn’t be possible to apply due there is no antibiotics left available with a corresponding backbone.
One option to get around this obstacle was to run the gel, after digestion of BB_I04060, and cut the band with the Biobrick-length and then 3A assembly it with the J-construct and E-P digested psB1C3.
Another option was to switch backbone of J-construct to psB1C3 and then use 3A-assembly it with BB_I0460 and E-P digested psB1C3. We used the second method because it’s a more safer way even though it takes longer time to finish.
 "
"