Team:Heidelberg/Modeling
From 2010.igem.org
(→Introduction into Fuzzy Logic) |
(→Fuzzy Inference Model) |
||
| Line 112: | Line 112: | ||
| - | + | How is our fuzzy inference system optimized? | |
| - | + | MISO Sugeno Fuzzy Network Model | |
| - | + | ||
| - | + | Optimizable | |
| - | + | ||
| - | + | Extendable | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===Fuzzy Model Concepts=== | ===Fuzzy Model Concepts=== | ||
| Line 144: | Line 130: | ||
Strength: general prediction, no dependency on conditions. Assured by [normalization strategy] | Strength: general prediction, no dependency on conditions. Assured by [normalization strategy] | ||
| + | |||
| + | based on previous knowledge [Bartel] | ||
| + | |||
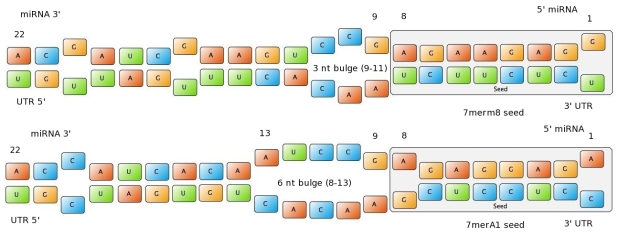
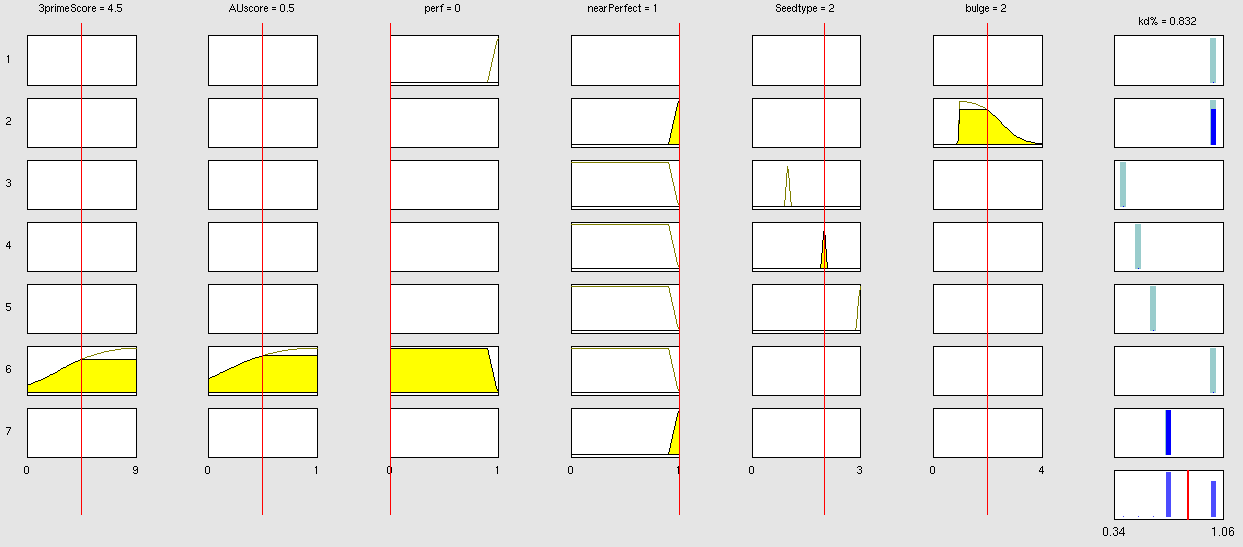
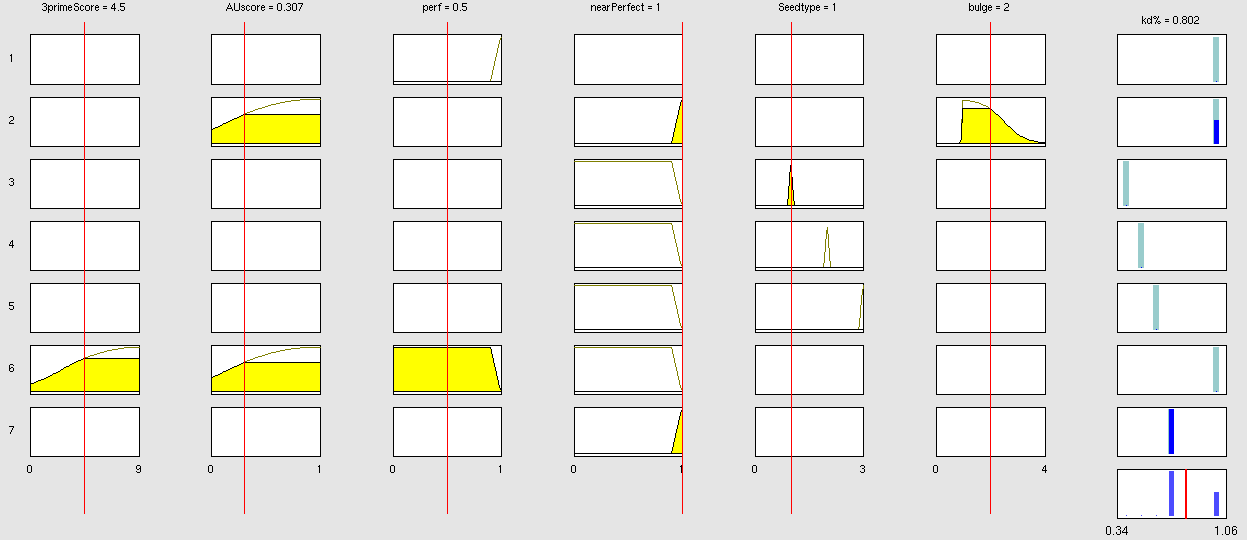
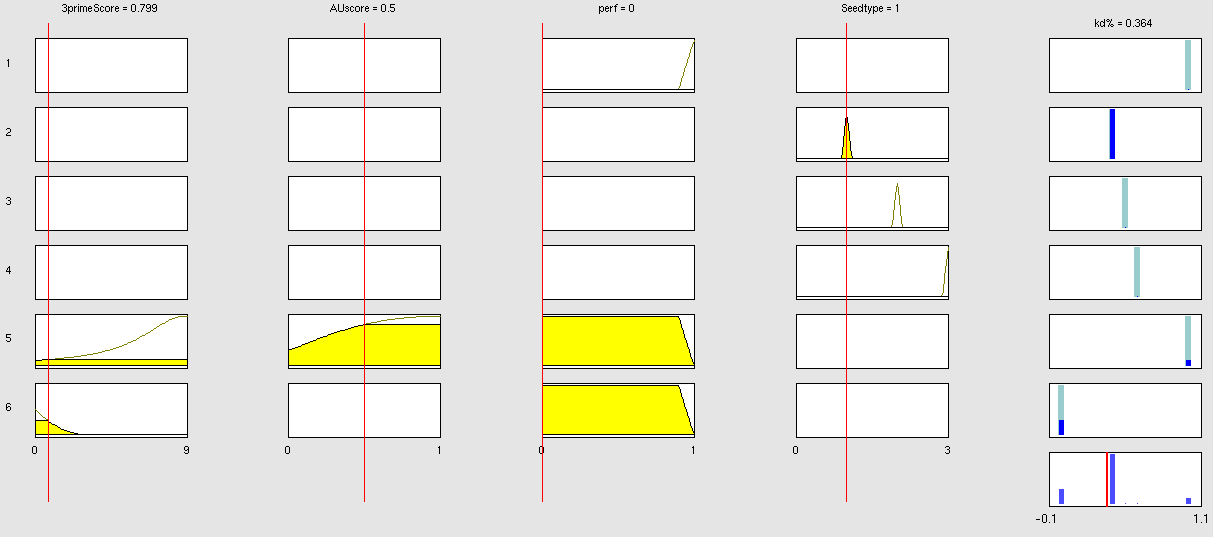
| + | Our fuzzy inference system can deal with 3 different kinds of shRNA binding sites. Perfect, bulged and endogenous-like binding sites are treated separately, due to the differences in their biological mechanism, as discussed earlier [link to binding site properties]. | ||
| + | A perfect binding site is evaluated by a simple ON/OFF input MF evaluating the boolean input of | ||
We came up with different concepts of what kind of input parameters to integrate into the fuzzy inference model and how to evaluate them. Therefore we parameterized the [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset properties of a large set of binding sites] according to various different BS characteristics. | We came up with different concepts of what kind of input parameters to integrate into the fuzzy inference model and how to evaluate them. Therefore we parameterized the [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset properties of a large set of binding sites] according to various different BS characteristics. | ||
The targetscan_50_context_scores – Algorithm {{HDref|Rodriguez et al., 2007}} which evaluates binding sites in respect to 3'-pairing and AU-content gives out a score that seems appropriate to distinguish especially between endogenous miRNA like binding sites. A more detailed description on the concept of binding site parameterization can be found under [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset Model Training Set]. | The targetscan_50_context_scores – Algorithm {{HDref|Rodriguez et al., 2007}} which evaluates binding sites in respect to 3'-pairing and AU-content gives out a score that seems appropriate to distinguish especially between endogenous miRNA like binding sites. A more detailed description on the concept of binding site parameterization can be found under [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset Model Training Set]. | ||
| + | |||
| + | Input parameters | ||
| + | |||
| + | Input membership functions | ||
| + | |||
| + | Output membership functions | ||
| + | |||
| + | Rules | ||
| + | |||
| + | |||
| + | Optimization | ||
| + | |||
| + | Parameters and their functionality | ||
| + | |||
| + | Output Membership function values | ||
| + | |||
| + | 7merA1 | ||
| + | |||
| + | 7merM8 | ||
| + | |||
| + | 8mer | ||
| + | |||
| + | (Nearperfect) | ||
| + | |||
| + | (Perfect) | ||
| + | |||
<html> | <html> | ||
| Line 154: | Line 171: | ||
</html> | </html> | ||
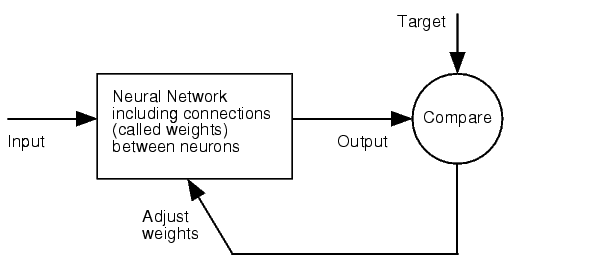
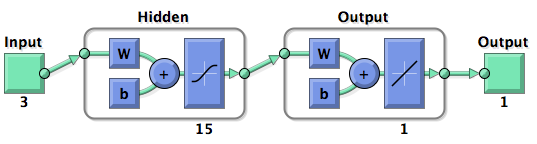
===Fuzzy Model Optimization=== | ===Fuzzy Model Optimization=== | ||
| + | |||
| + | |||
| + | |||
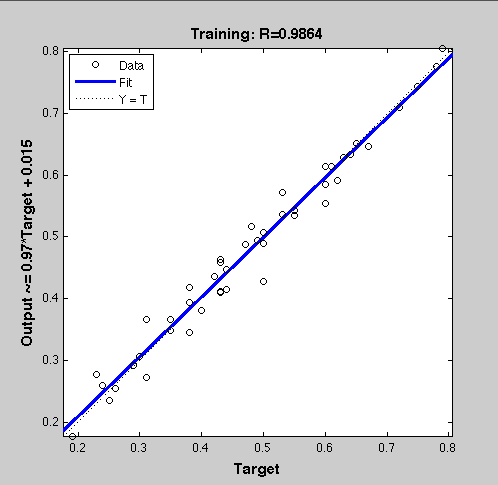
| + | ===Result=== | ||
[http://igem.bioquant.uni-heidelberg.de/igem_2010/FuzzyModelResults.html Click here, if you are interested in more recent model optimizations results!] | [http://igem.bioquant.uni-heidelberg.de/igem_2010/FuzzyModelResults.html Click here, if you are interested in more recent model optimizations results!] | ||
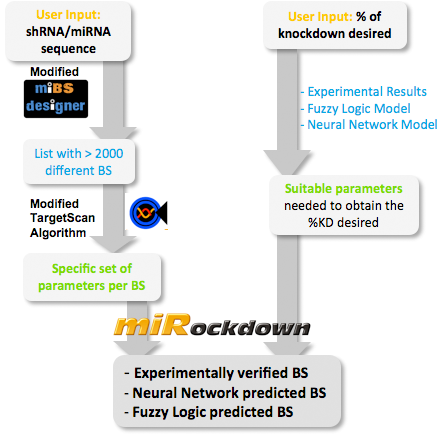
| - | + | ==miRockdown on miBEAT== | |
Right from the beginning of our modeling project, we knew we would have to integrate our trained models into an online GUI. We realized it in the most user friendly way we could think of: The user only needs to input the desired knockdown percentage (kd%) and choose an sh/miRNA sequence, to get a binding site that satisfies the users needs.<br> | Right from the beginning of our modeling project, we knew we would have to integrate our trained models into an online GUI. We realized it in the most user friendly way we could think of: The user only needs to input the desired knockdown percentage (kd%) and choose an sh/miRNA sequence, to get a binding site that satisfies the users needs.<br> | ||
<br> | <br> | ||
Revision as of 19:34, 26 October 2010

|
|
|||
 "
"