Team:Tsinghua/experiments
From 2010.igem.org
| Line 137: | Line 137: | ||
[[Image:THU-hdy-1.JPG]] | [[Image:THU-hdy-1.JPG]] | ||
| + | |||
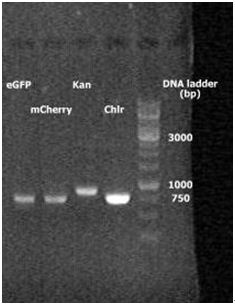
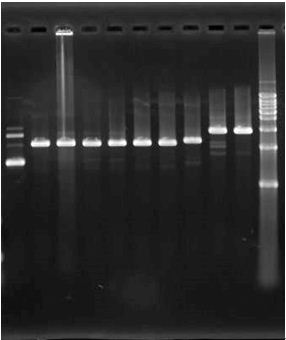
2. 2010.8.16 RCR result for eGFP, mCherry, Kan and Chlr. | 2. 2010.8.16 RCR result for eGFP, mCherry, Kan and Chlr. | ||
| - | [[Image:THU-hdy-2.JPG]] | + | [[Image:THU-hdy-2.JPG|200px]] |
| + | |||
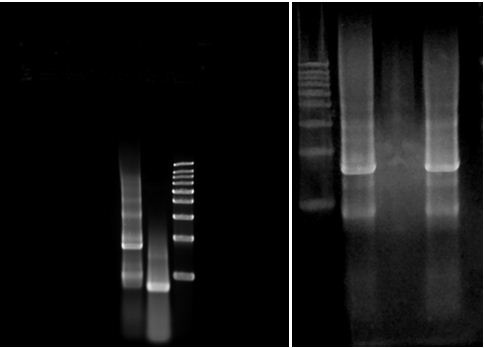
3. Result : plasmid digestion with EcoR I and Pst I. | 3. Result : plasmid digestion with EcoR I and Pst I. | ||
[[Image:THU-hdy-3.JPG]] | [[Image:THU-hdy-3.JPG]] | ||
| + | |||
4. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, there was no positive clone. | 4. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, there was no positive clone. | ||
| - | [[Image:THU-hdy-4.JPG]] | + | [[Image:THU-hdy-4.JPG|200px]] |
| + | |||
5. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, two clones, whose size is about 3Kb (plasmid+ Chlr), are positive. | 5. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, two clones, whose size is about 3Kb (plasmid+ Chlr), are positive. | ||
| - | [[Image:THU-hdy-5.JPG]] | + | [[Image:THU-hdy-5.JPG|300px]] |
| + | |||
6. As no positive clone for plasmid-eGFP-mCherry-Kan is picked, we have strong doubt about the efficiency of ligation of multiple fragments. Thus, PCR is conducted to confirm whether the ligation reaction took place in the system. Upstream primer of eGFP and downstream primer of Kan are used in PCR. Bands with the size of about 2.5Kb is hardly seen in the gel photograph. | 6. As no positive clone for plasmid-eGFP-mCherry-Kan is picked, we have strong doubt about the efficiency of ligation of multiple fragments. Thus, PCR is conducted to confirm whether the ligation reaction took place in the system. Upstream primer of eGFP and downstream primer of Kan are used in PCR. Bands with the size of about 2.5Kb is hardly seen in the gel photograph. | ||
| - | [[Image:THU-hdy-6.JPG]] | + | [[Image:THU-hdy-6.JPG|300px]] |
<html> | <html> | ||
Revision as of 16:36, 26 October 2010

Experiment Records
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Our Experiments were carried out by the following nine groups which are somewhat independent from each other. Here come the Groups and Their Tasks.
Group 1A:Landing pad (LP) construction and Insertion Project with Att Recomination method and Trandition Landing pad
Protocols
Here are our Experimental Protocols in PDF version. You're advised to view in Acrobat or Adobe Reader which you can download free from the Home Website of Adobe® Company, for other viewers we can not guarantee the right format of viewing. Those files are encrypted to keep copyrights, if you want to print them out of copy to your documents, to share with others our ideas and methods, you are welcome to ask us for the unencrypted files definitely for free~Molecular Cloning
THUProtocol_1-1_Isolatio_of_plasmid_DNA
THUProtocol_1-2_DNA_Gel_Extraction
THUProtocol_1-3_Restriction_Enzyme_Digestion
THUProtocol_1-4_Ligation_of_DNA_Fragments
THUProtocol_1-5_PCR
THUProtocol_1-6_Preparation_of_Competent_Cell_for_Electro_Transformation
THUProtocol_1-7_Chemical_Transformation_of_Recombinant_DNA
THUProtocol_1-8_Electro_Transformation_of_Recombinant_DNA
Protein Isolation and Identification
THUProtocol_2-1_Protein_Isolation_for_Prokaryotes
THUProtocol_2-2_Protein_Identification_SDS-PAGE
THUProtocol_1-5_PCR
Results
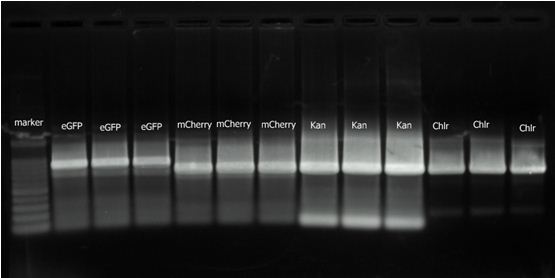
Group 2(b)1. PCR: Amplication eGFP, mCherry, Kanamycin resistant gene and Chlr four genes separatly from PIB-eGFP, PBS34, PKD13 and PKD3 plasmids.
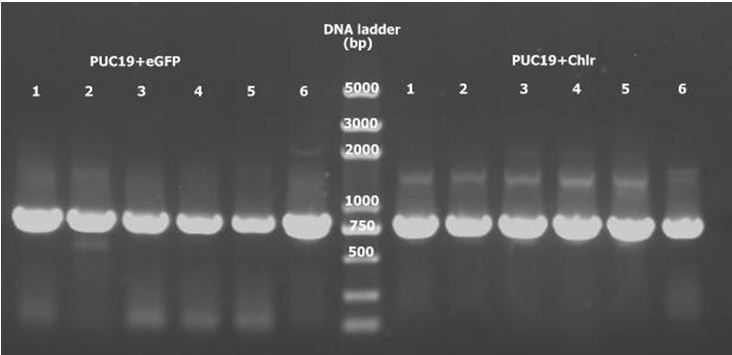
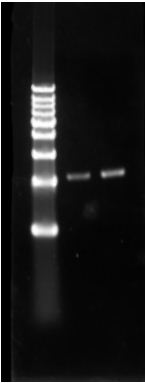
2. pUC19 double digestion for eGFP and Chlr, ligate with digested eGFP and Chlr separately. PCR to detect pUC19+eGFP and pUC19+ChlrPCR. All 12 samples are positive.
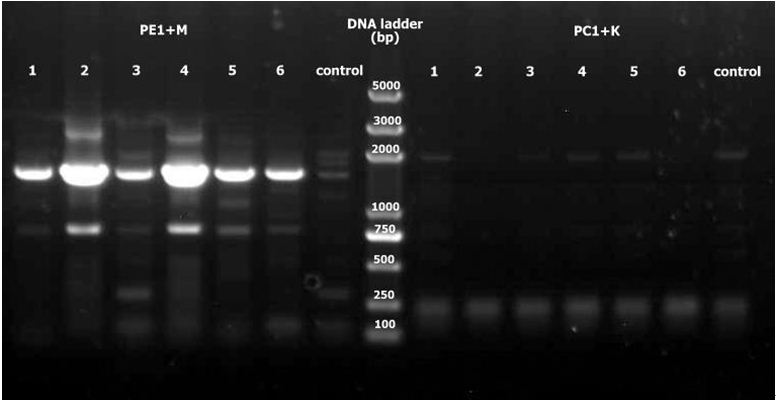
3. Double digest PE1 and mCherry with SalI and BamHI, double digest PC1 and Kan with KpnI and BamHI, ligate and PCR to detect. No positive results exist among PC1+K.
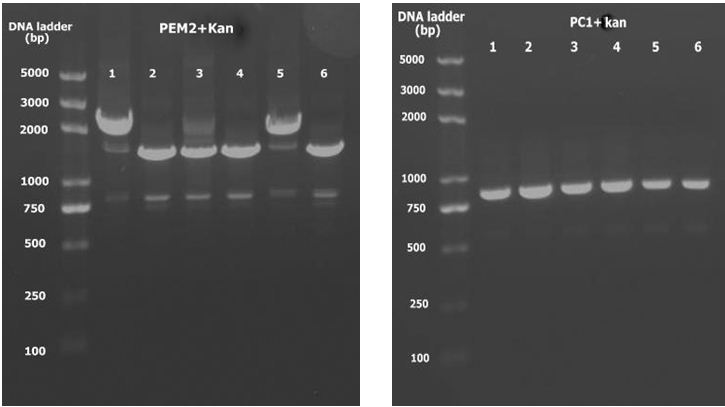
4. Double digest PEM2 and Kan with KpnI and BamHI as well as PC1. Ligate and using PCR to detect with universal primers. PEM2+K 1 and 5 are positive.
5. Double digest PEMK1 and Chlr with KpnI and EcoRI. Ligate and using PCR to detect with universal primers. PEMK1+Chlr 3-5 are positive.
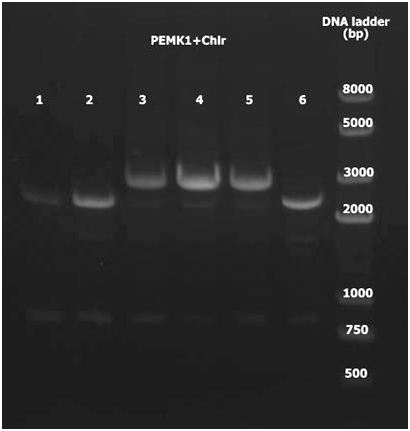
Group 2(c)
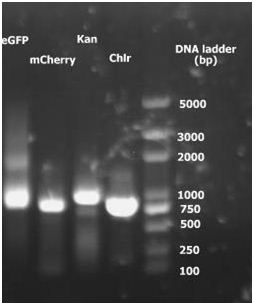
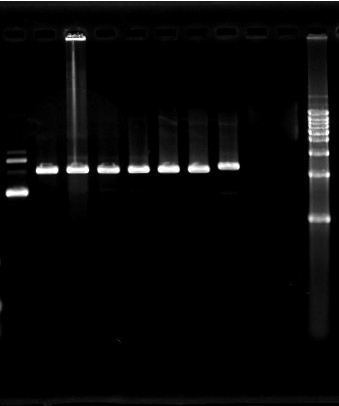
1. 2010.7.31 PCR result for eGFP, Kan and Chlr. No specific target bands appeared in the photograph. Besides, dimerization of primers is severe.
2. 2010.8.16 RCR result for eGFP, mCherry, Kan and Chlr.
3. Result : plasmid digestion with EcoR I and Pst I.
4. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, there was no positive clone.
5. Identification: After digestion, ligation and transformation, plasmid is extracted and digested with EcoR I. The size of positive clone is about 2.7Kb while the negative is less than 2Kb. As the photograph shows, two clones, whose size is about 3Kb (plasmid+ Chlr), are positive.
6. As no positive clone for plasmid-eGFP-mCherry-Kan is picked, we have strong doubt about the efficiency of ligation of multiple fragments. Thus, PCR is conducted to confirm whether the ligation reaction took place in the system. Upstream primer of eGFP and downstream primer of Kan are used in PCR. Bands with the size of about 2.5Kb is hardly seen in the gel photograph.
 "
"