Team:Newcastle/Filamentous Cells
From 2010.igem.org
Swoodhouse (Talk | contribs) (→Research) |
Swoodhouse (Talk | contribs) (→Characterisation) |
||
| Line 54: | Line 54: | ||
==Characterisation== | ==Characterisation== | ||
| - | ==== | + | ===Selection for integration=== |
| - | + | ||
| - | + | ||
To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be | To select for integration of the plasmid into the chromosome, ''B. subtilis'' will be | ||
| Line 69: | Line 67: | ||
The construct we designed has a GFP transcriptional fusion after the ''yneA'' coding sequence, so GFP is co-transcribed and acts as our fluorescent marker for transcription of ''yneA''. | The construct we designed has a GFP transcriptional fusion after the ''yneA'' coding sequence, so GFP is co-transcribed and acts as our fluorescent marker for transcription of ''yneA''. | ||
| - | + | ===Lab work and Results=== | |
We characterised the part first without, and then with, LacI repression (using the integration vector pMutin4 to integrate ''lacI'' into the ''Bacillus subtilis'' 168 chromosome. | We characterised the part first without, and then with, LacI repression (using the integration vector pMutin4 to integrate ''lacI'' into the ''Bacillus subtilis'' 168 chromosome. | ||
Revision as of 16:31, 26 October 2010

| |||||||||||||
| |||||||||||||
Contents |
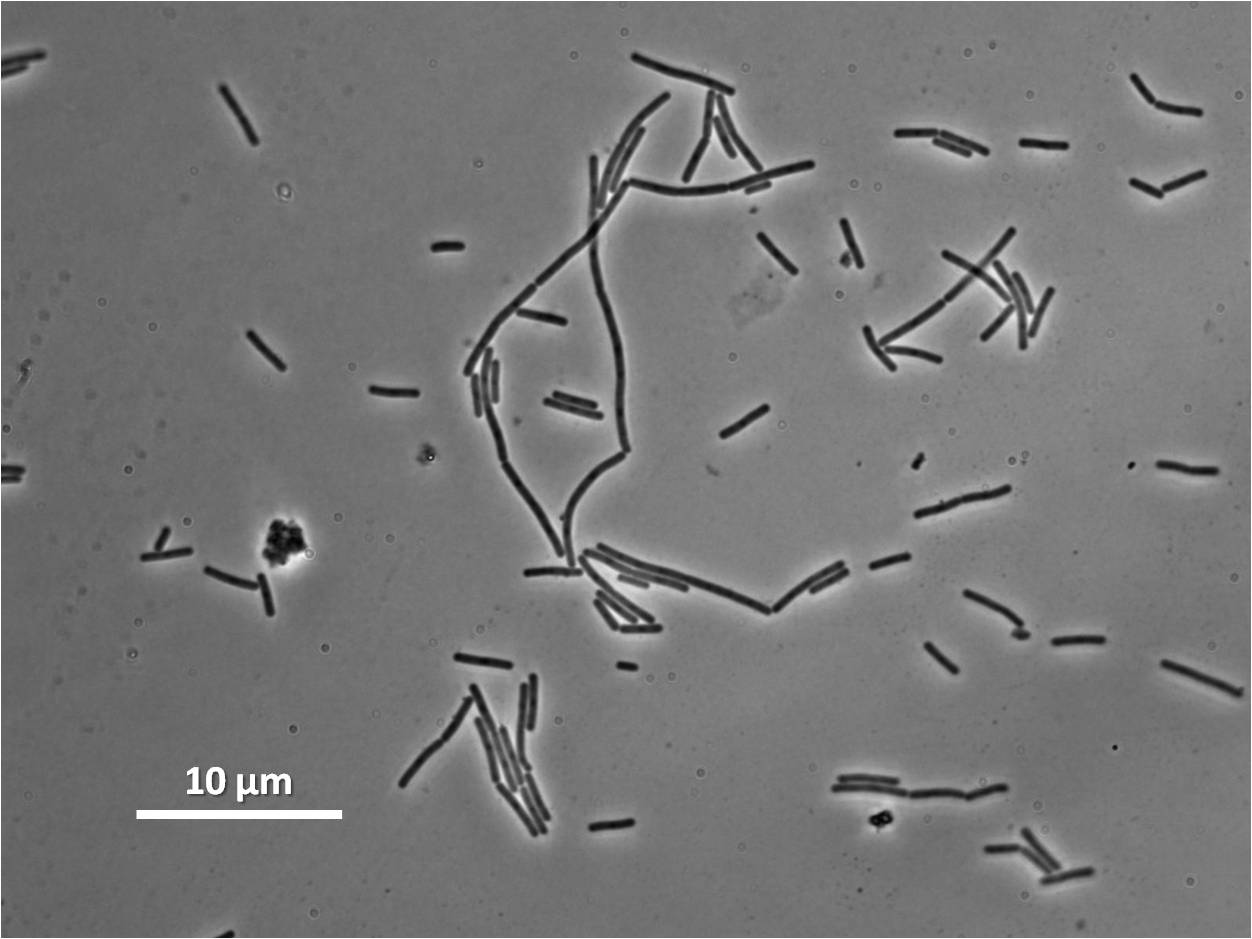
Filamentous cell formation by overexpression of yneA
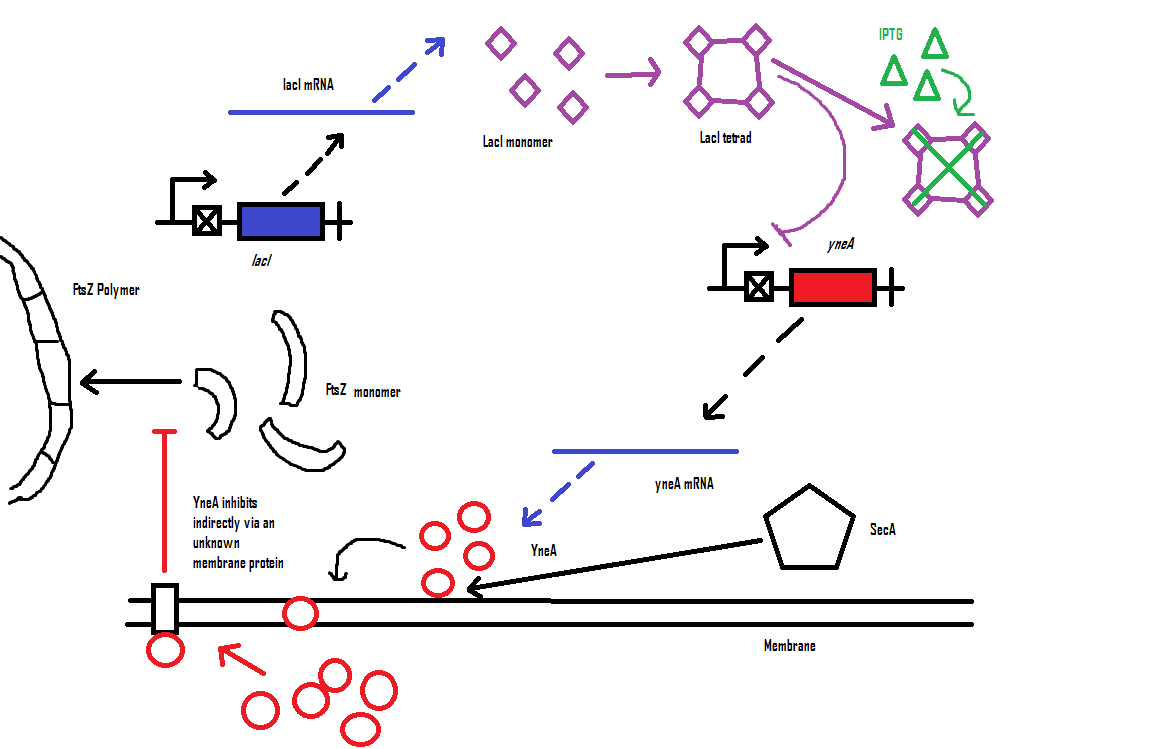
Bacillus subtilis cell division is dependent on FtsZ. FtsZ forms a 30 subunit ring at the midpoint of the cell and contracts.
YneA indirectly stops the formation of the FtsZ ring. In nature, yneA is expressed during SOS response, allowing the cell to repair DNA damage before continuing with the division cycle.
It is hypothesized that YneA acts through unknown transmembrane proteins to inhibit FtsZ ring formation; we call these unknown components "Blackbox proteins".
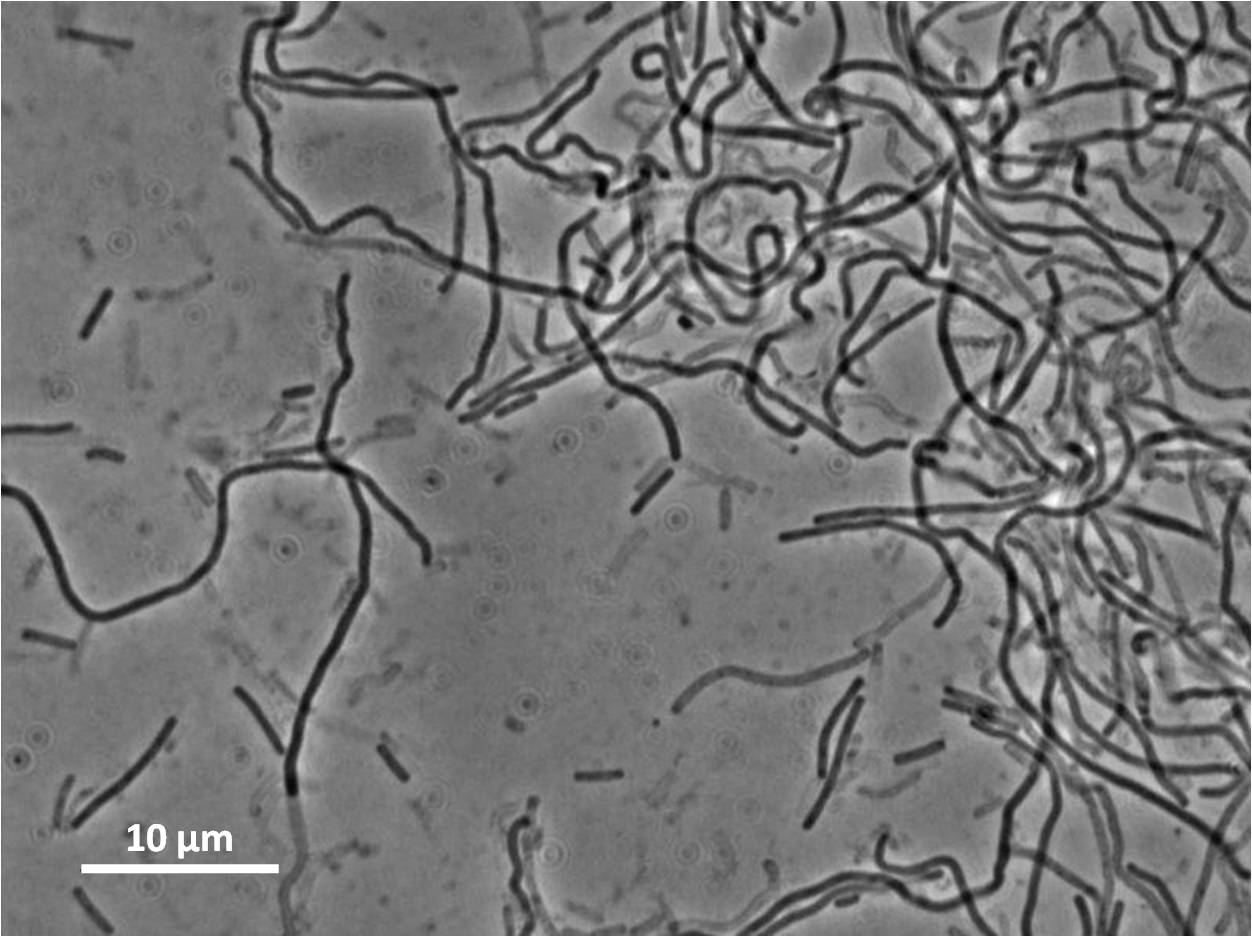
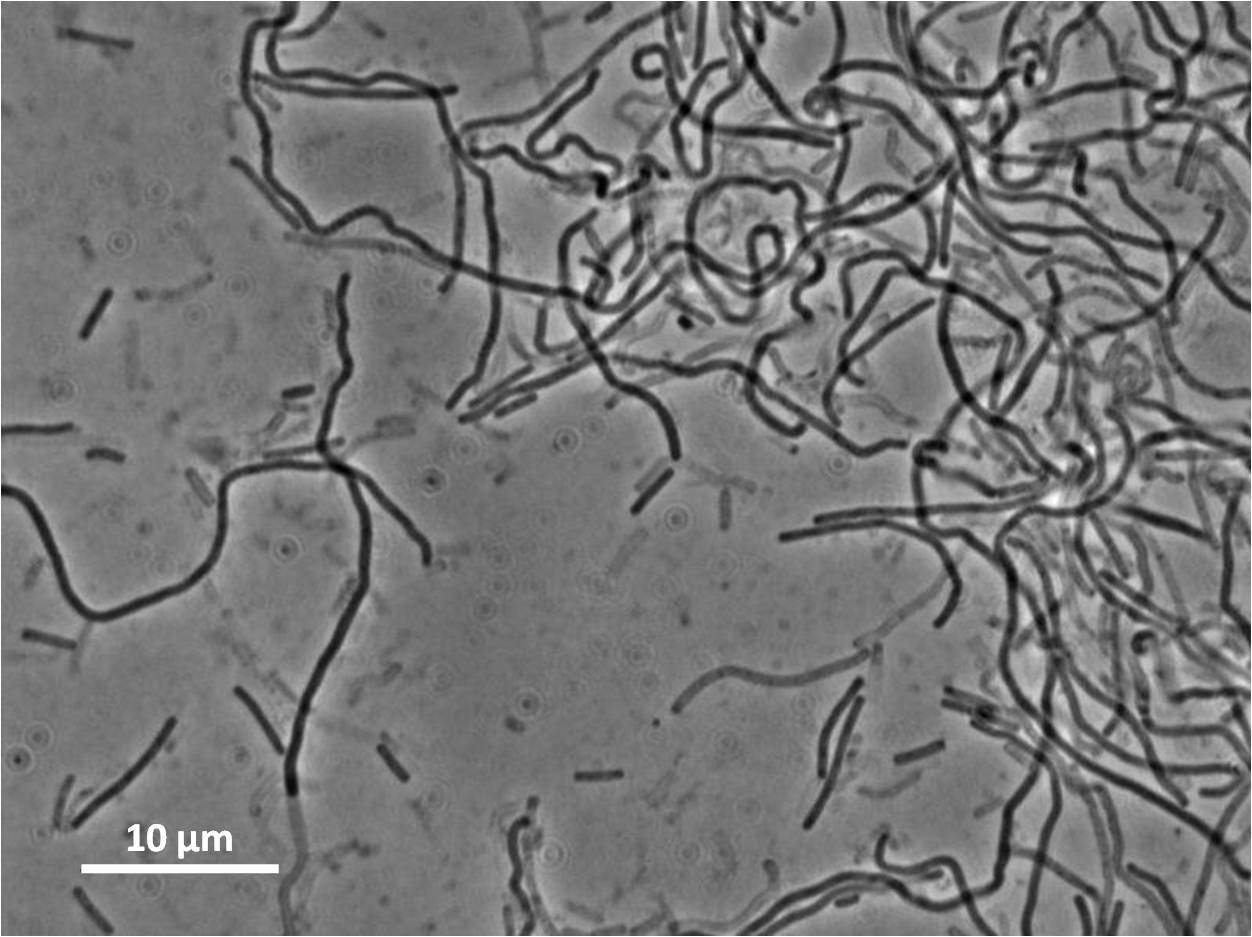
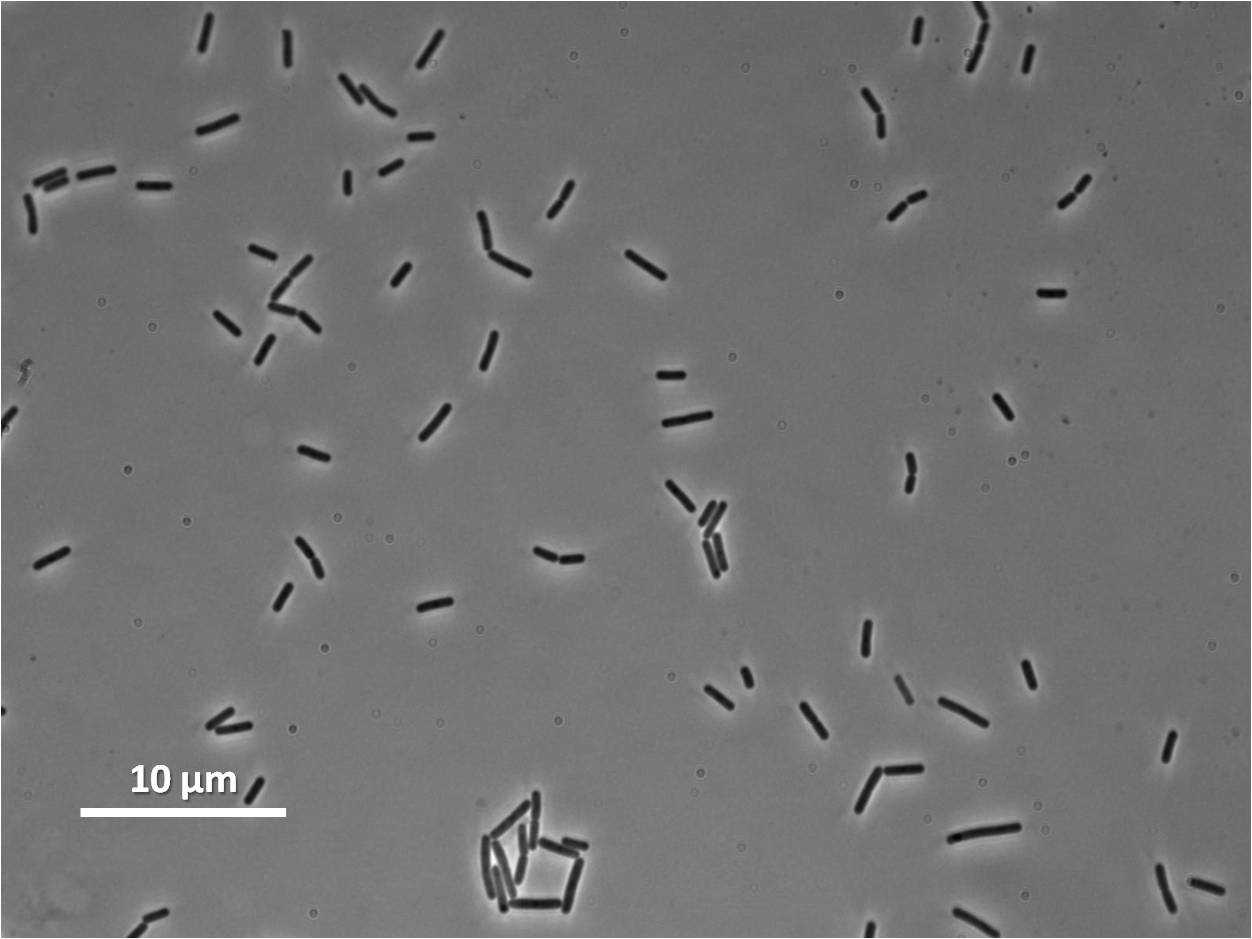
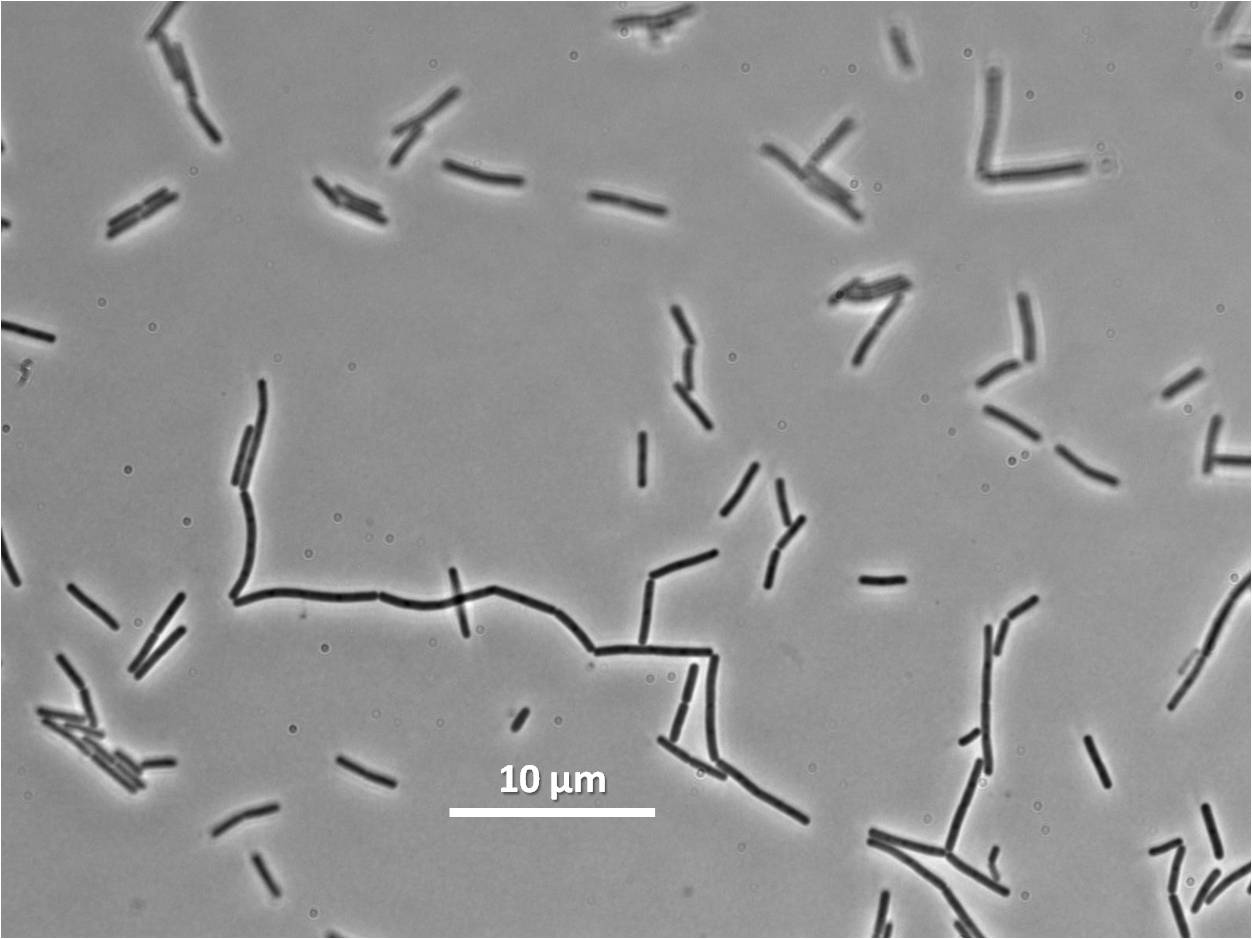
By expressing YneA and therefore inhibiting FtsZ ring formation, the cells will grow filamentous.
Part
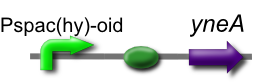
Our IPTG-inducible filamentous cell formation part puts yneA under the control of the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K302003 strongly LacI-repressible promoter that we designed, hyperspankoid]. In the presence of LacI, induction with IPTG will result in a filamentous cell phenotype.
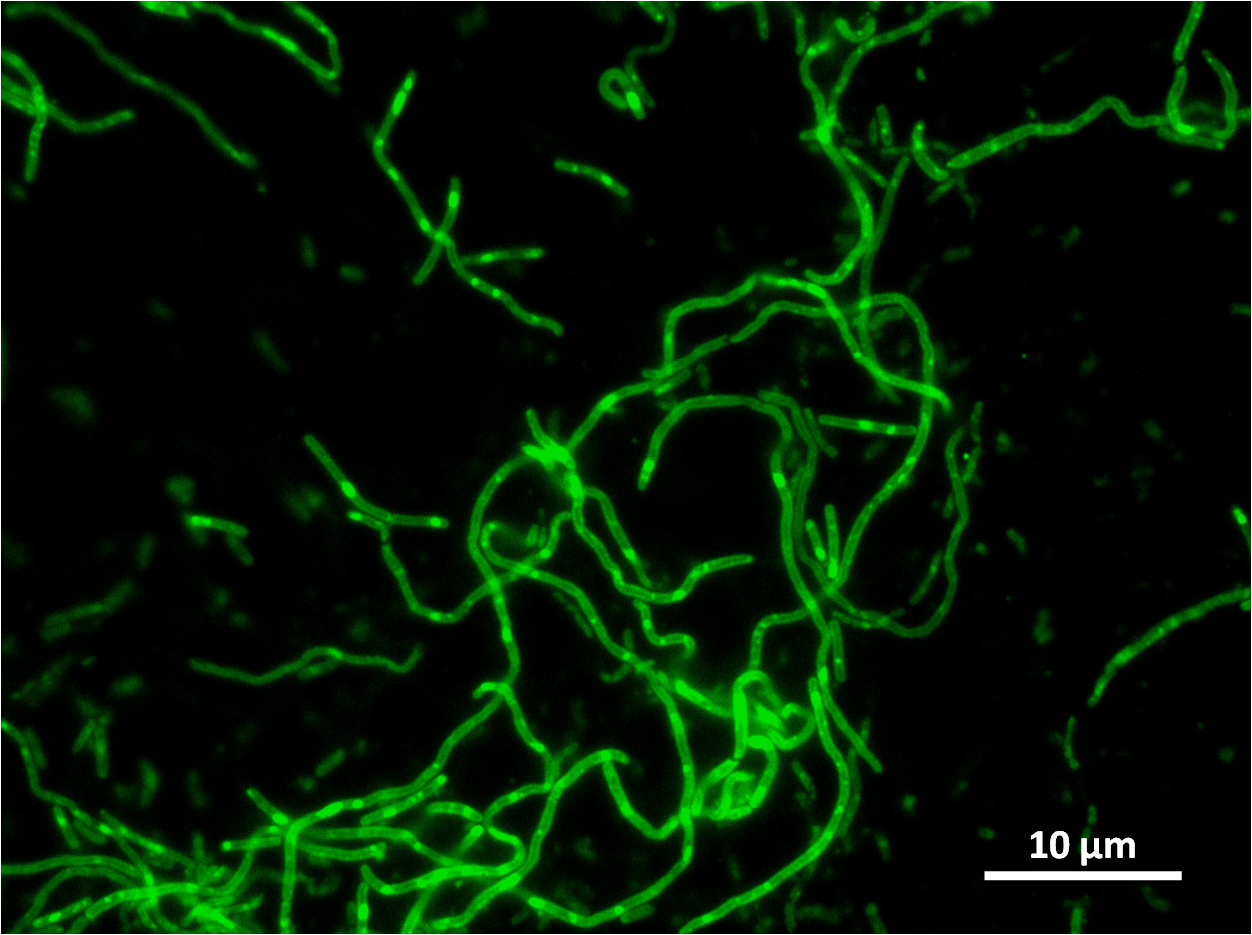
The part has no terminator, allowing for transcriptional fusion with gfp and visualisation under the microscope.
This is part [http://partsregistry.org/Part:BBa_K302012 BBa_K302012] on the [http://partsregistry.org parts registry].
Computational model
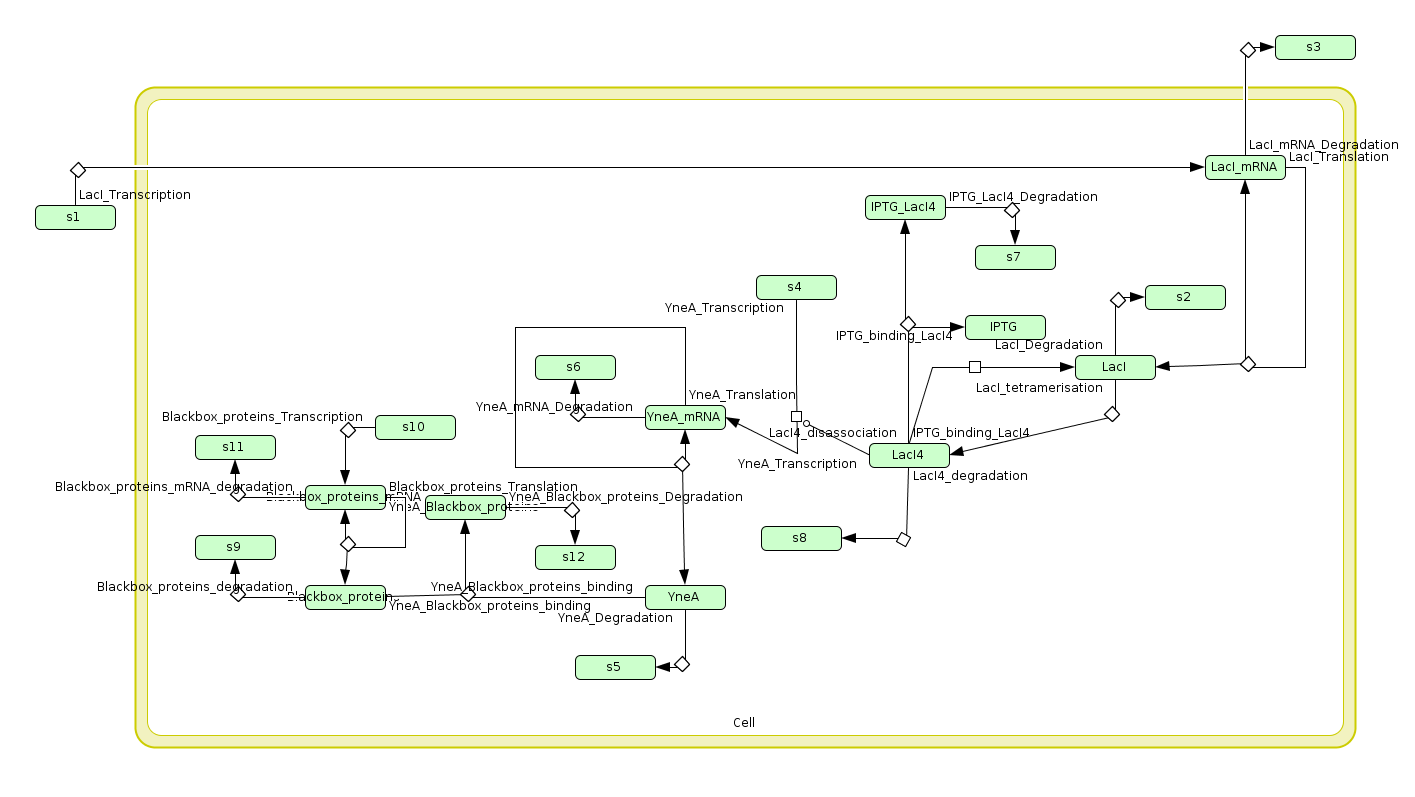
| Visualisation of the model's biochemical network in CellDesigner. |
Downloads:
Cloning strategy
Characterisation
Selection for integration
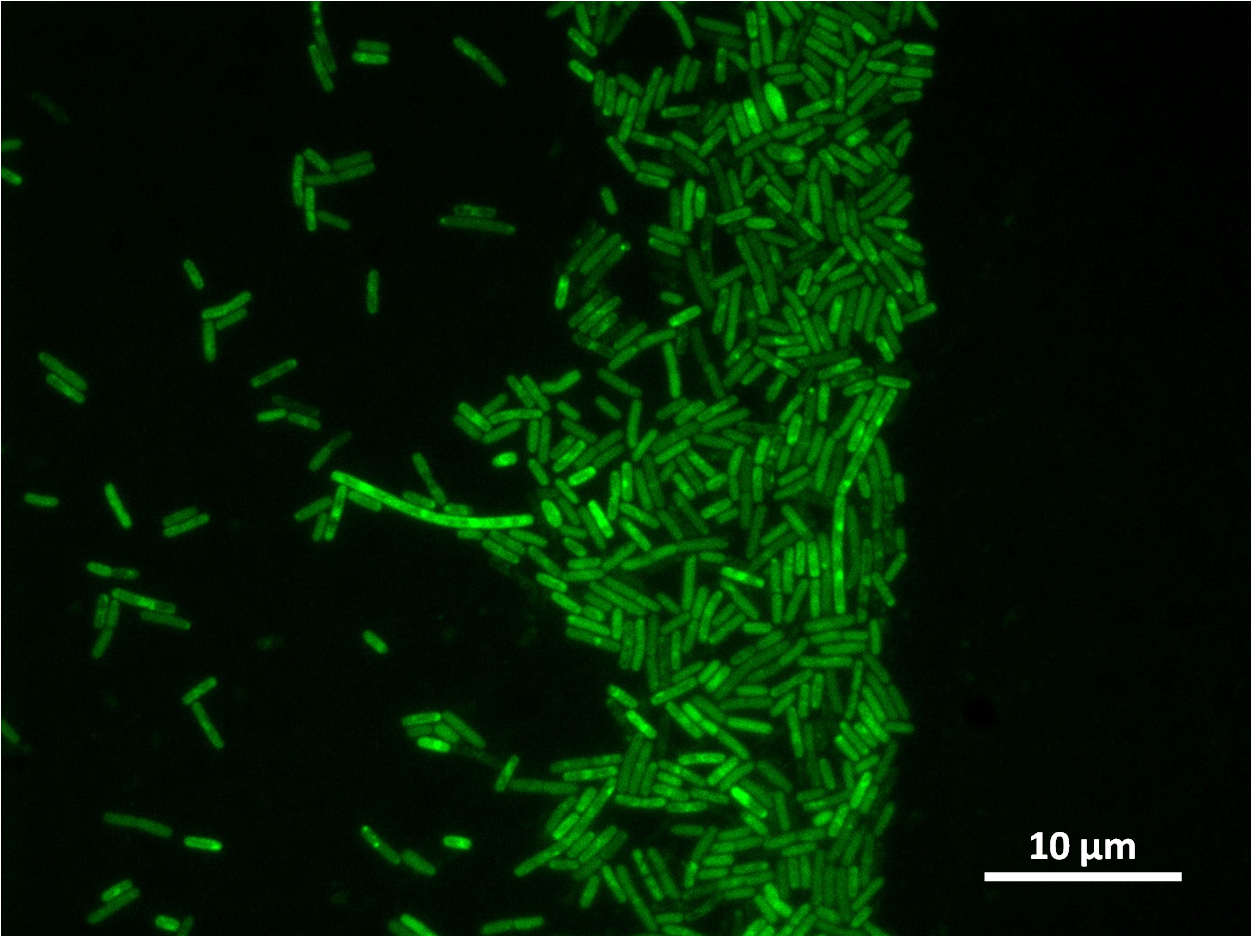
To select for integration of the plasmid into the chromosome, B. subtilis will be tested for the ability to hydrolyse starch. Integration of the BioBrick will be done by homologous recombination at the amyE, therefore destroying endogenous expression of amylase. Colonies that are not able to break down starch on agar plate will be selected and cultured for further test. Colonies that do not contain the integrated BioBrick will be able to hydrolyse starch, therefore forming a white halo around the colony as iodine interacts with starch to form blue colour.
The construct we designed has a GFP transcriptional fusion after the yneA coding sequence, so GFP is co-transcribed and acts as our fluorescent marker for transcription of yneA.
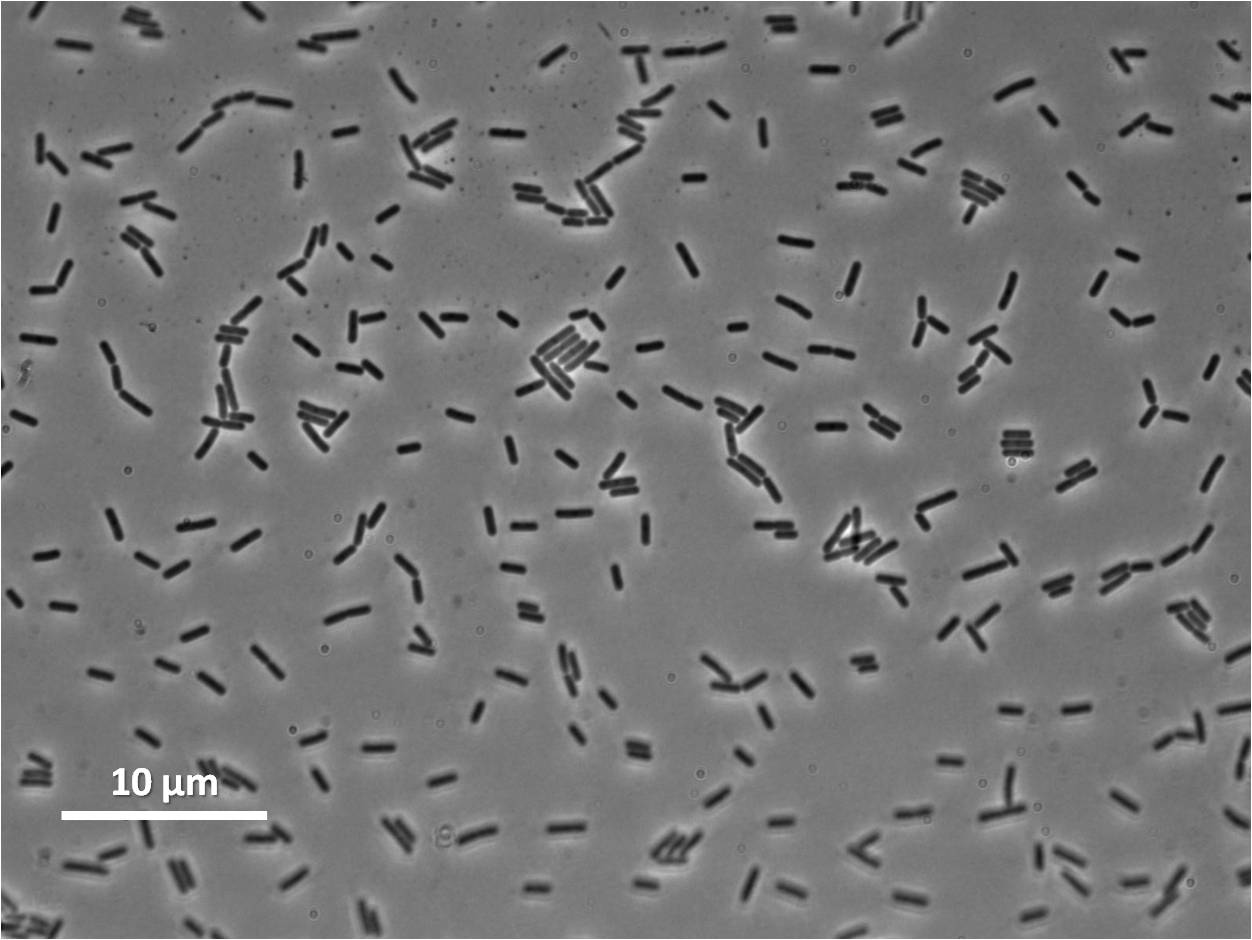
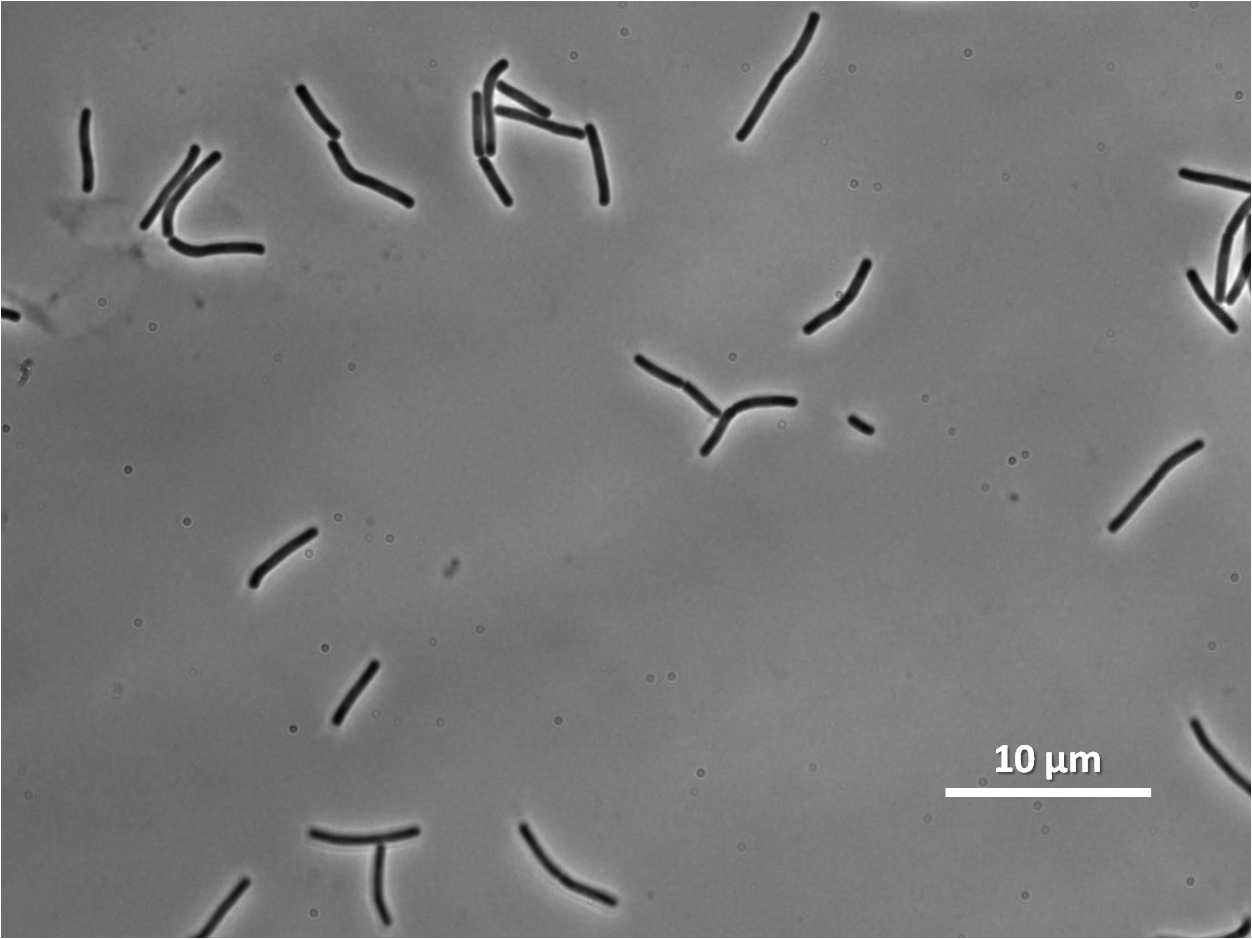
Lab work and Results
We characterised the part first without, and then with, LacI repression (using the integration vector pMutin4 to integrate lacI into the Bacillus subtilis 168 chromosome.
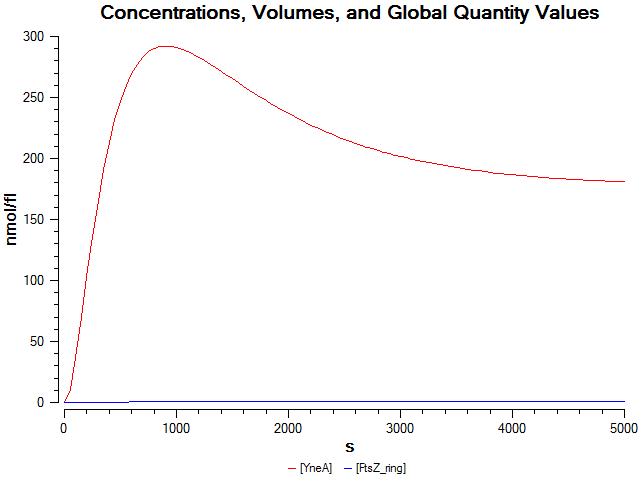
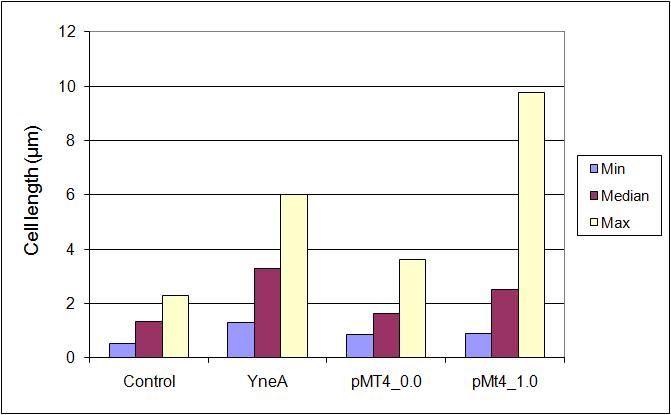
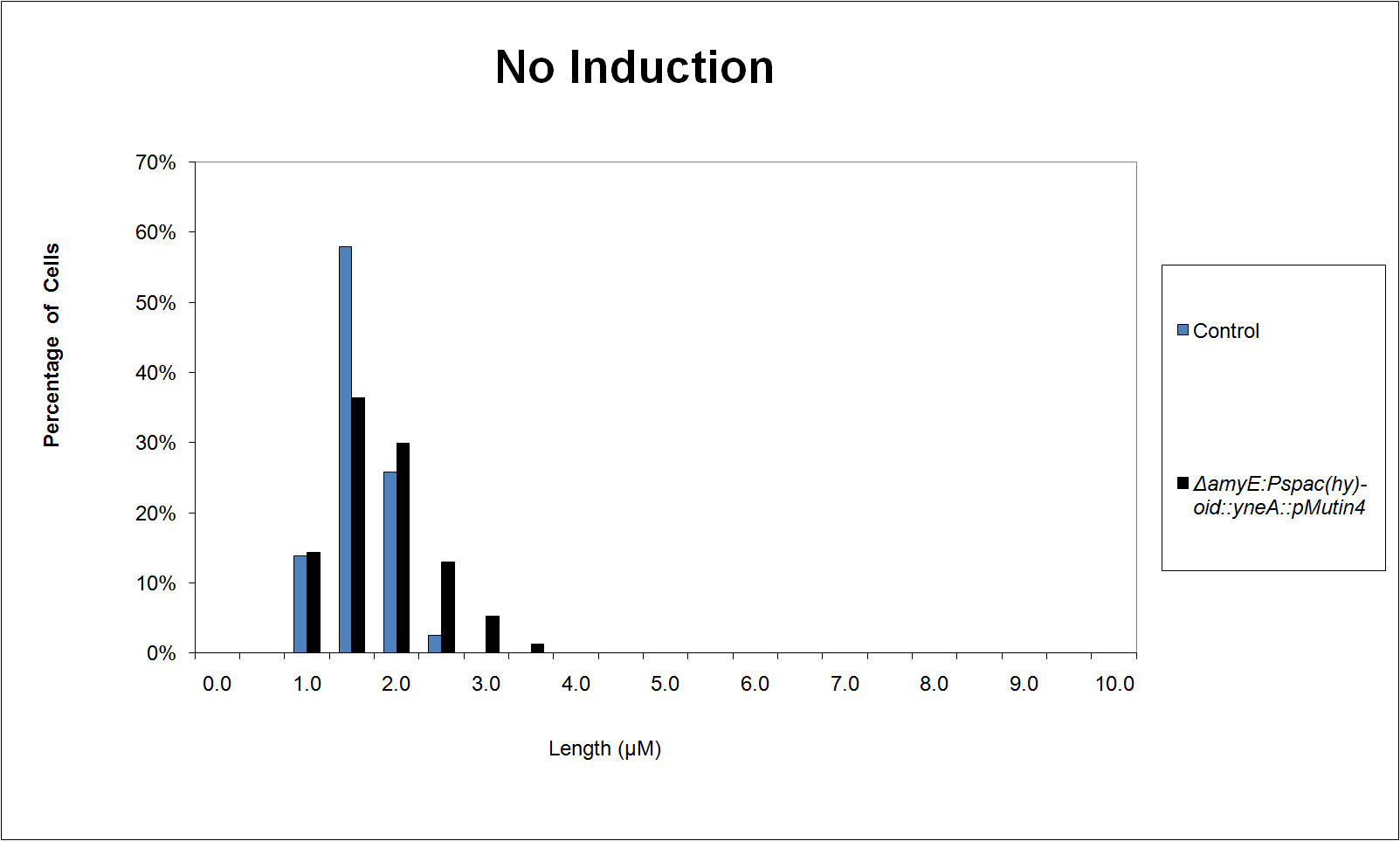
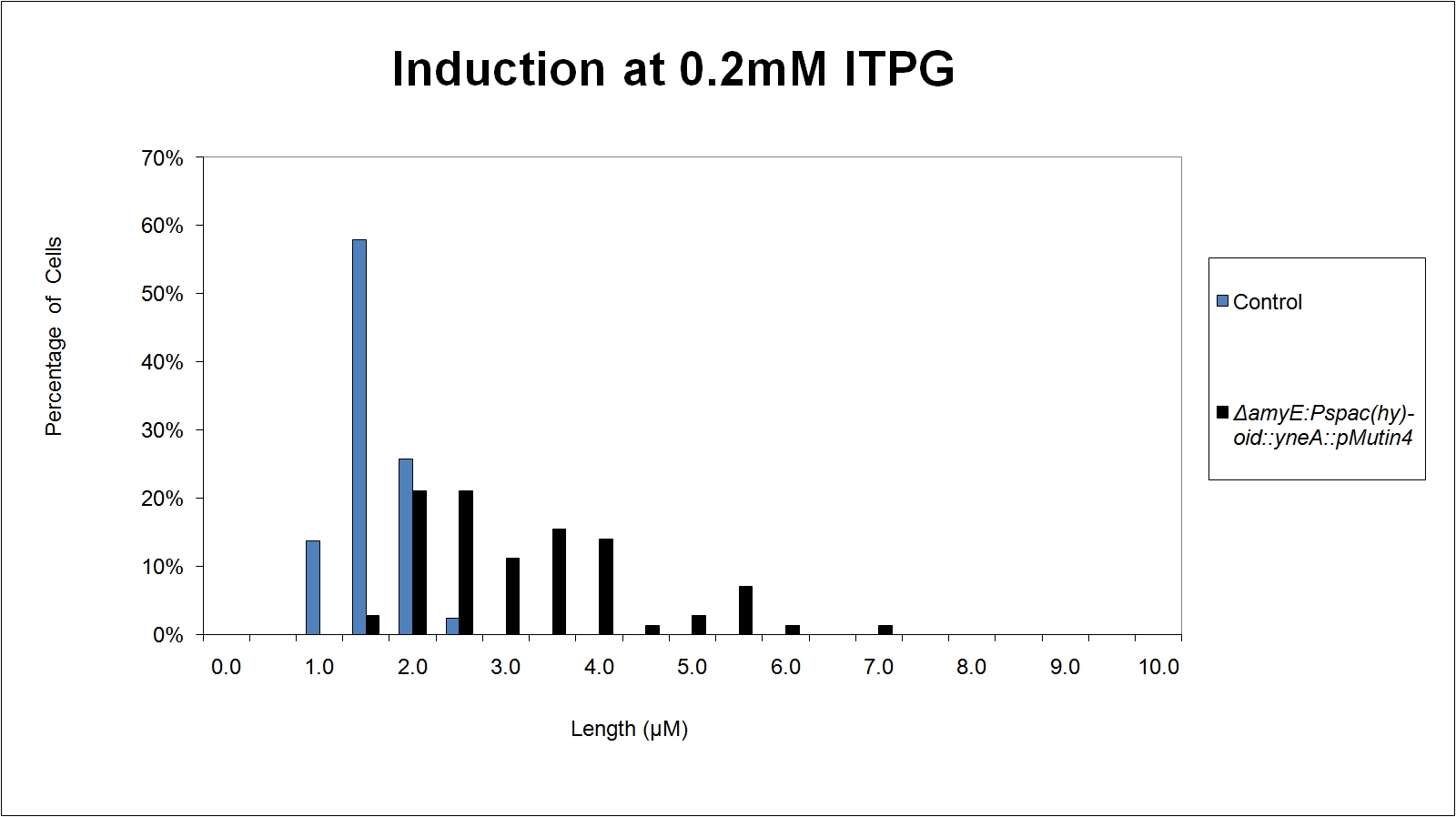
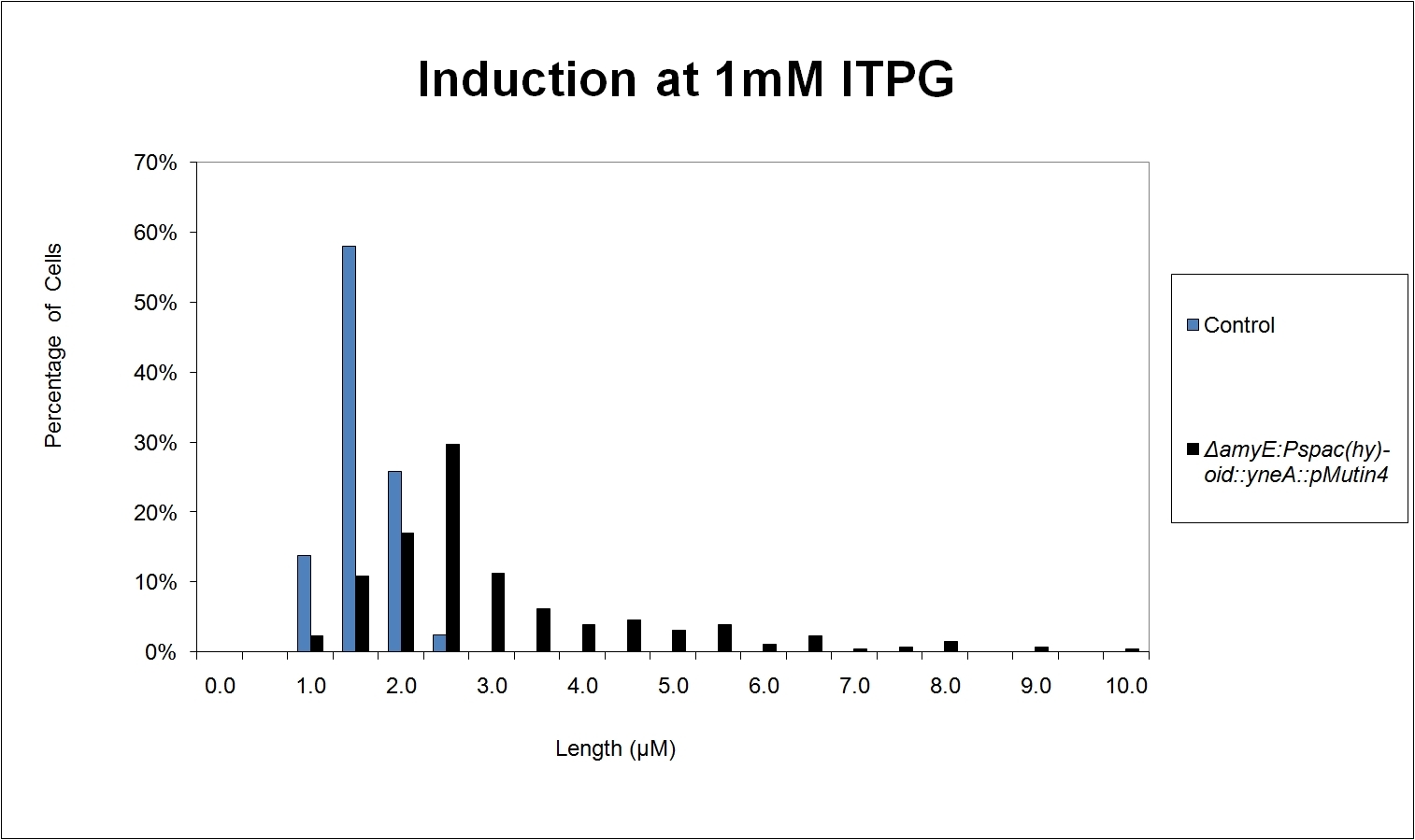
Graphs
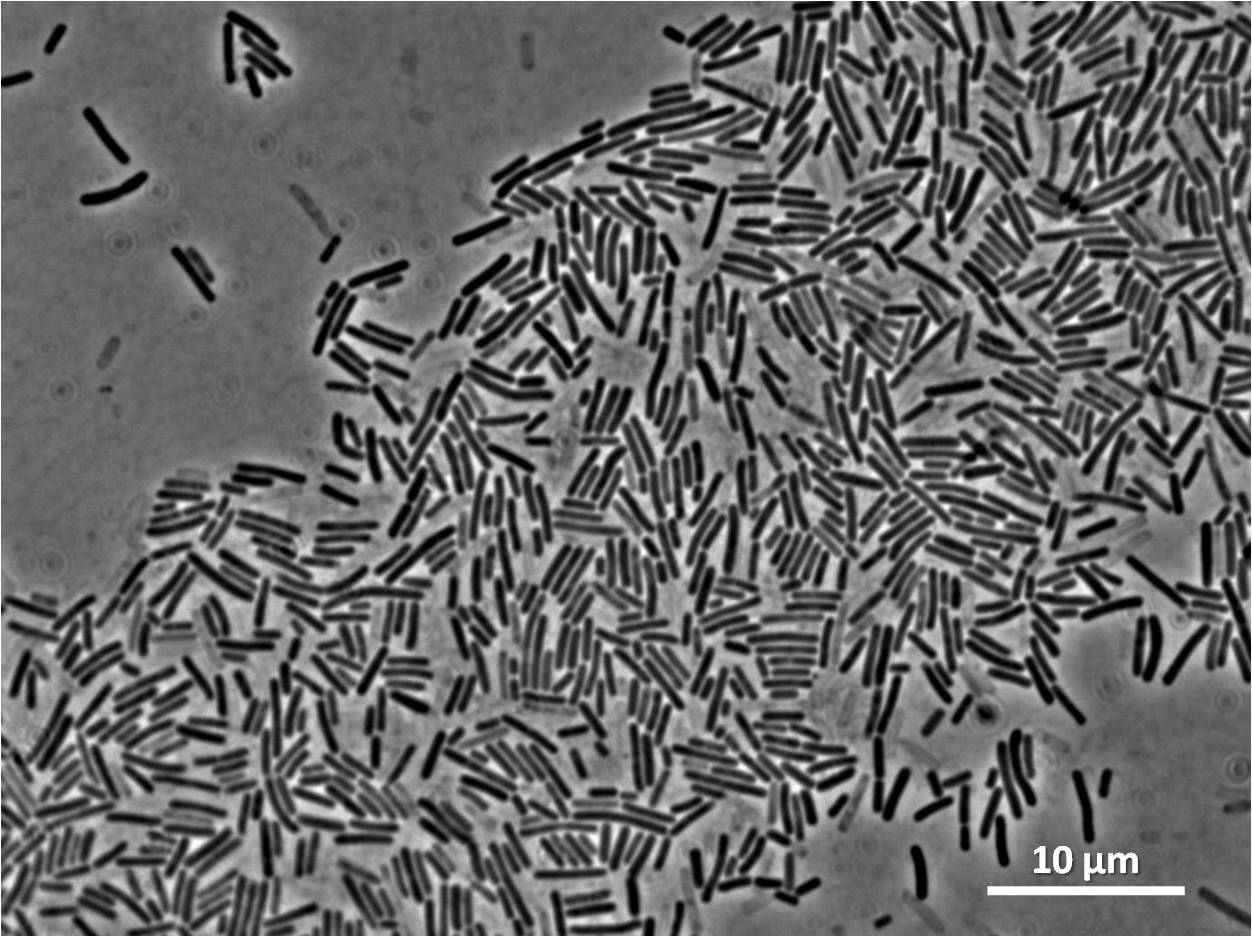
Graphs 2, 3 and 4 show a greater proportion of filamentous cells at a higher concentration of IPTG(1mM IPTG), compared with Bacillus subtilis 168 our control population.
Research
References
Kawai, Y., Moriya, S., & Ogasawara, N. (2003). "Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis". Molecular microbiology, 47(4), 1113-22.
Mo, A.H. & Burkholder, W.F., (2010). "YneA , an SOS-Induced Inhibitor of Cell Division in Bacillus subtilis , Is Regulated Posttranslationally and Requires the Transmembrane Region for Activity" ᰔ †. Society, 192(12), 3159-3173.
 
|
 "
"