Team:NYU/Parts
From 2010.igem.org
| Line 297: | Line 297: | ||
1. To improve parts already in the registry by lessening the constrictions on their use and making them more adaptable. | 1. To improve parts already in the registry by lessening the constrictions on their use and making them more adaptable. | ||
| + | [[Image:NYU_Cre.jpg|300px|right]] | ||
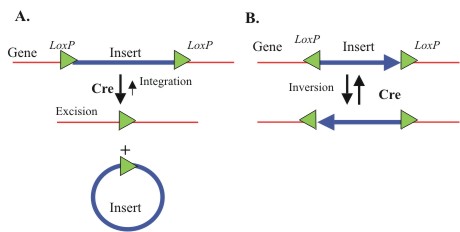
*'''36bp LoxP site''': the LoxP site is the naturally-occurring substrate for the Cre recombinase. In nature (and in previous biobricks) it is 34 base pairs long. Because it is not a multiple of three, when used inside a transcribed ORF the 34bp LoxP site can throw off the frame of the translated parts. To get around this, previous teams had to either add or take away bases from surrounding parts to keep their systems in frame (painstakingly and sometimes unsuccessfully). To get around this restriction, we designed and Biobricked a 36bp LoxP site that Registry users can include in transcribed regions without worrying about keeping everything in frame. | *'''36bp LoxP site''': the LoxP site is the naturally-occurring substrate for the Cre recombinase. In nature (and in previous biobricks) it is 34 base pairs long. Because it is not a multiple of three, when used inside a transcribed ORF the 34bp LoxP site can throw off the frame of the translated parts. To get around this, previous teams had to either add or take away bases from surrounding parts to keep their systems in frame (painstakingly and sometimes unsuccessfully). To get around this restriction, we designed and Biobricked a 36bp LoxP site that Registry users can include in transcribed regions without worrying about keeping everything in frame. | ||
Revision as of 06:18, 26 October 2010

When choosing what simple parts to make for our project we had two main goals: 1) to improve the quality of parts already in the registry by making them more accessible to all users and 2) to augment the current set of yeast-compatible biobricks to harness the power of this synthetic biology-friendly model organism. Incorporating these goals with the 'Quality not Quantity' mantra, we decided to focus our efforts on certain areas of advancement that we believe future researchers will like to see in the registry.
1. To improve parts already in the registry by lessening the constrictions on their use and making them more adaptable.
- 36bp LoxP site: the LoxP site is the naturally-occurring substrate for the Cre recombinase. In nature (and in previous biobricks) it is 34 base pairs long. Because it is not a multiple of three, when used inside a transcribed ORF the 34bp LoxP site can throw off the frame of the translated parts. To get around this, previous teams had to either add or take away bases from surrounding parts to keep their systems in frame (painstakingly and sometimes unsuccessfully). To get around this restriction, we designed and Biobricked a 36bp LoxP site that Registry users can include in transcribed regions without worrying about keeping everything in frame.
- pGAL-RBS:
- pTEF-RBS:
- lexOpCYC-RBS:
2. Yeast strains are more rarely used in iGEM projects but contain certain parameters that are ideal for certain projects. Many teams are beginning to use yeast this year and are submitting useful biobricks, so we decided to make a collection of biobricks parts aimed at protein localization within the cell (known as 'yeast functional tags'):
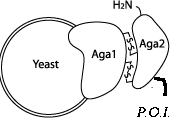
- Aga2: When the Aga2 protein is fused to a part of interest, the fusion is secreted and Aga2 complexes with a protein on the cellular surface - leaving the part of interest displayed on the surface of the cell. While we did not end up using this part in our official construct, biobricking it from a plasmid we already had in our lab will surely benefit future teams.
- Aga2+Linker: Most everyone will want a linker attached to the Aga2 protein, so we went ahead and submitted this construct as well.
- Secretion Tag: Another 'yeast functional tag', the secretion tag does just what you think when fused to the C terminus of a protein of interest.
- Ura3:
3. To construct and submit certain ubiquitously useful biobricks that we would be using.
- Flexible (Gly4Ser)3 peptide linker: for use with most any peptide fusions
Parts
New for iGEM 2010 is the groupparts tag. This tag will generate a table with all of the parts that your team adds to your team sandbox. Note that if you want to document a part you need to document it on the [http://partsregistry.org Registry], not on your team wiki.
<groupparts>iGEM010 NYU</groupparts>
 "
"