Team:Newcastle/31 August 2010
From 2010.igem.org
(→Gibson cloning of the rocF BioBrick) |
(→Second transformation of 'Bacillius subtilis 168' with Prrnb-GFP containing YneA) |
||
| Line 119: | Line 119: | ||
| - | =Second transformation of 'Bacillius subtilis | + | =Second transformation of ''Bacillius subtilis'' 168 with Prrnb-GFP containing YneA= |
==Aim== | ==Aim== | ||
| - | The aim of the experiment is to perform insert the plasmid Prrnb-GFP containing YneA which have been ligated eariler into the chromosome of ''Bacillus subtilis | + | The aim of the experiment is to perform insert the plasmid Prrnb-GFP containing YneA which have been ligated eariler into the chromosome of ''Bacillus subtilis'' 168. ''B. subtilis'' containing the intergated vector will be resistance to both the antibiotic chloramphenicol and streptomycin, therefore those that have successful intergated will be selected with agar plates that contain these antibiotic. The second step will be to identify those colones that have the plasmid intergated at the corerct position in the chromosome, which is the amylase locus. Thus those that have intergrated at the wrong position will not be able to break down starch, which can be tested with the iodine test. |
==Materials and Protocol== | ==Materials and Protocol== | ||
Please refer to: [[Team:Newcastle/Transformation of B. subtilis| Transformation of ''Bacillus subtilis'']] | Please refer to: [[Team:Newcastle/Transformation of B. subtilis| Transformation of ''Bacillus subtilis'']] | ||
| - | Note: Overnigth culture of ''B. subtilis | + | Note: Overnigth culture of ''B. subtilis'' 168 in MM competence medium was done the day before and the iodine test was performed the day after. |
==Results== | ==Results== | ||
Revision as of 03:38, 26 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Amplification of the circular plasmid pSB1C3 for the Subtilin immunity BioBrick
Aim
The aim of this experiment is to amplify the plasmid pSB1C3 for the construction of the subtilin immunity BioBrick with the help of two Phusion PCR reactions using old and new primers, and to perform gel electrophoresis to check whether amplification was successful.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol and gel electrophoresis. The details for the PCR reactions are given below.
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | pSB1C3 | P1V1 forward | P2V1 reverse | 58 | 2072 approx. | 65 |
| 2 | Plasmid Vector | pSB1C3 | pSB1C3_for | pSB1C3_rev | 68 | 2072 approx. | 65 |
Table 1: Table represents two Phusion PCR reactions amplifying the plasmid pSB1C3 so that it can be ligated together with other Subtilin Immunity fragments using the Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Therefore the extension time of each PCR reaction is different.
Discussion
After gel electrophoresis the correct band was found with the original primers, but no band was found with the new primers. We will proceed to gel extraction of the correct band tomorrow.
Amplification of the parts for Subtilin Immunity BioBrick
Aim
The aim of this experiment is to amplify the four parts for the Subtilin Immunity BioBrick using Phusion PCR again but using the original primers that were ordered and hydrated. (Before this could be done, four of the primers, 1-T1_for, 2-T1_rev, 1-P1_for and 2-P2_rev, had to be re-hydrated.)
For more information about the subtilin immunity BioBrick, please see the cloning strategy for subtilin immunity (Link)
Materials and Protocol
Please refer to Phusion PCR for Phusion PCR protocol. (Please refer to DNA Re-hydration for DNA Re-hydration protocol.)
The details for the four PCR reactions are included in the table below:
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Plasmid Vector | linear PSB1C3 (cut with HindIII) | P1V1 forward | P2V1 reverse | 53.3 (53) | 2046 + | 70 |
| 2 | Promoter and RBS (pVeg-SpoVG) | BioBrick Bba_K143053 | P1P1 forward | P2P2 reverse | 51.7 (51) | 139 + | 15 |
| 3 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 53.0 (53) | 2753 + | 110 |
| 4 | Double terminator | pSB1AK3 consisting BBa_B0014 | P1T1 forward | P2T1 reverse | 50.9 (51) | 116 + | 15 |
Table 2: This table shows the four different Phusion PCR reactions that were carried out today. If this is successful, the four parts will be ligated together for the construction of subtilin immunity BioBrick with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1 Kb/ 30 seconds. Thus the extension time of each and every PCR reaction is slightly different.
Results, Discussion and Conclusion
Gel electrophoresis and Gel extraction will be carried out if the bands are successful.
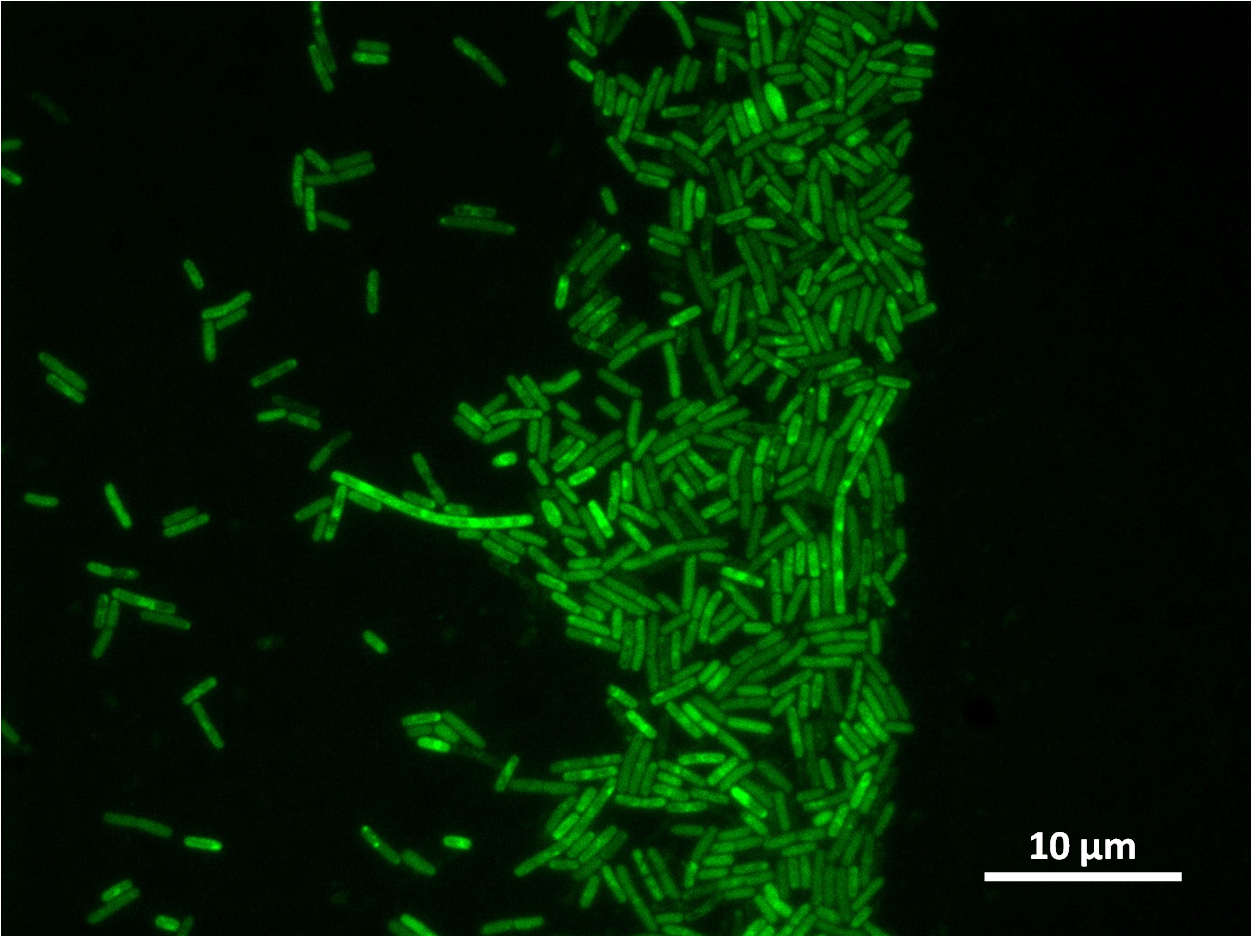
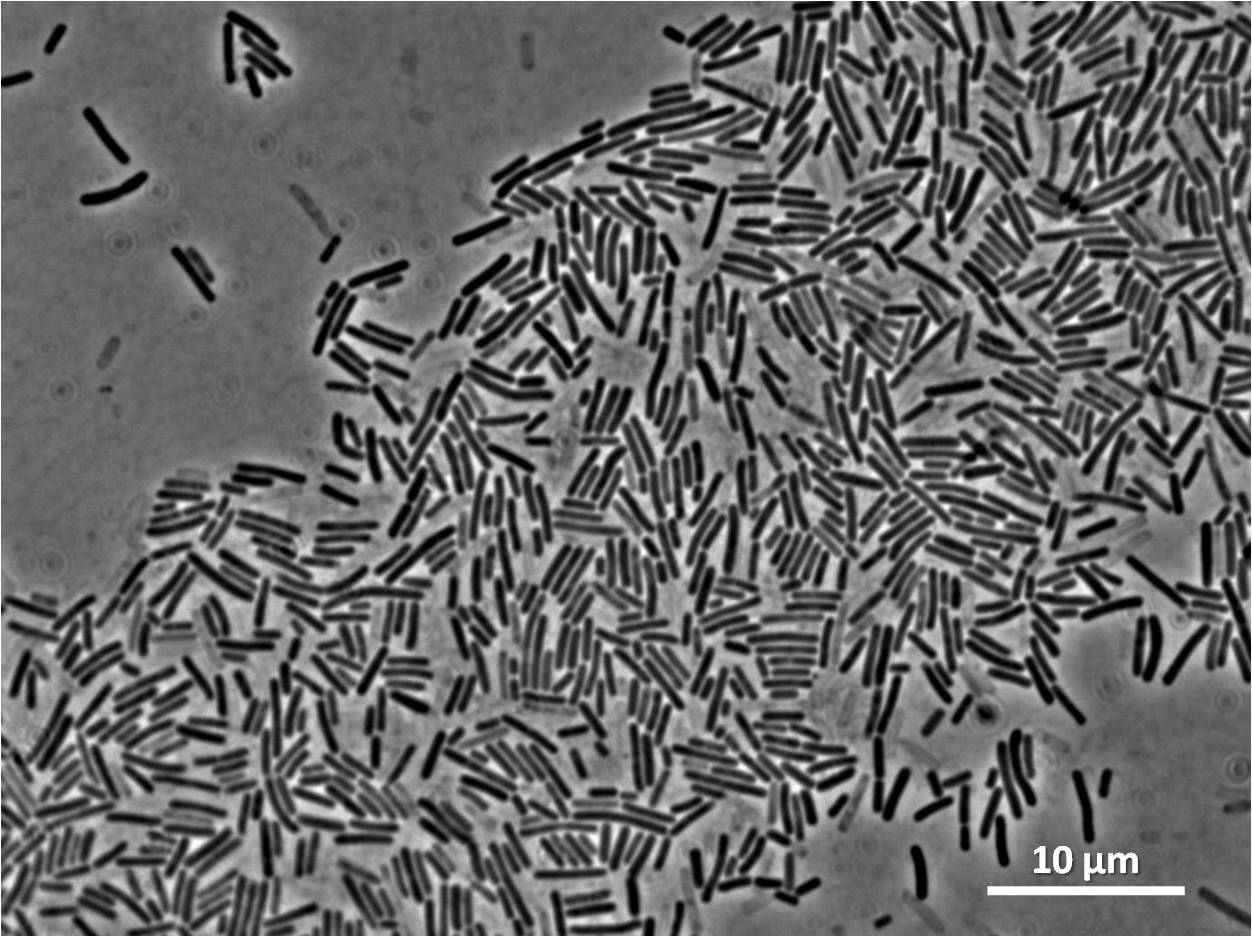
Second transformation of Bacillius subtilis 168 with Prrnb-GFP containing YneA
Aim
The aim of the experiment is to perform insert the plasmid Prrnb-GFP containing YneA which have been ligated eariler into the chromosome of Bacillus subtilis 168. B. subtilis containing the intergated vector will be resistance to both the antibiotic chloramphenicol and streptomycin, therefore those that have successful intergated will be selected with agar plates that contain these antibiotic. The second step will be to identify those colones that have the plasmid intergated at the corerct position in the chromosome, which is the amylase locus. Thus those that have intergrated at the wrong position will not be able to break down starch, which can be tested with the iodine test.
Materials and Protocol
Please refer to: Transformation of Bacillus subtilis Note: Overnigth culture of B. subtilis 168 in MM competence medium was done the day before and the iodine test was performed the day after.
Results
 
|
 "
"